Epidemiology of Classic and Novel Human Astrovirus: Gastroenteritis and Beyond
Abstract
:1. Introduction
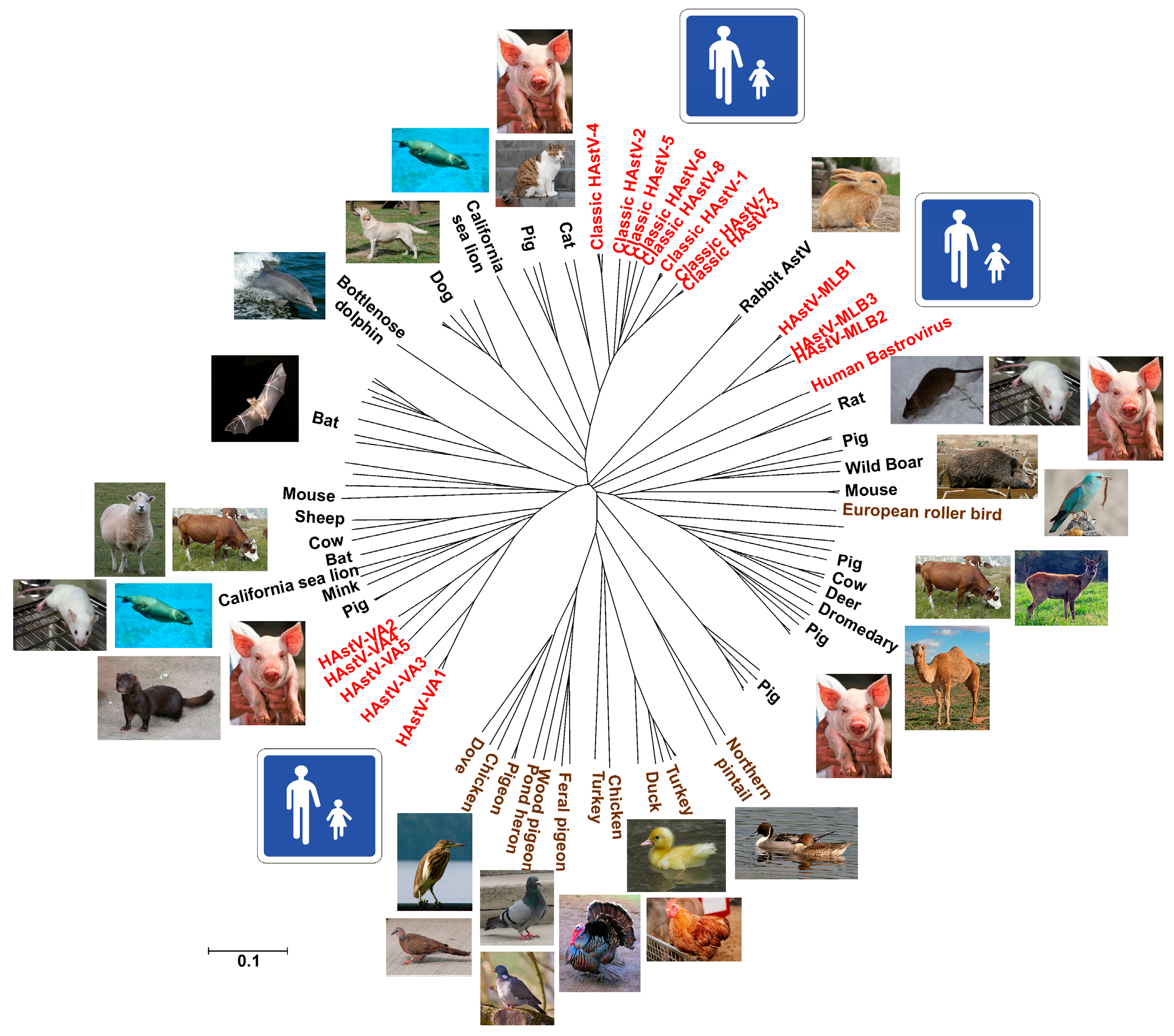
2. Classification
3. Diversity and Zoonotic Potential
4. Pathogenesis and Disease Spectrum
4.1. Astrovirus in the Gastrointestinal Tract
4.2. Astrovirus beyond the Gastrointestinal Tract and in Other Organs
5. Prevalence and Distribution
5.1. Classic HAstVs
5.2. Major Reported Outbreaks
5.3. Novel Astroviruses
6. Transmission Routes
7. Control and Prevention
8. Conclusions
Acknowledgments
Conflicts of Interest
References
- Appleton, H.; Higgins, P.G. Letter: Viruses and gastroenteritis in infants. Lancet 1975, 1, 1297. [Google Scholar] [CrossRef]
- Bosch, A.; Pinto, R.M.; Guix, S. Human astroviruses. Clin. Microbiol. Rev. 2014, 27, 1048–1074. [Google Scholar] [CrossRef] [PubMed]
- Mendez, E.; Arias, C.F. Astroviruses. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2007; Volume 5, pp. 981–1000. [Google Scholar]
- Jarchow-Macdonald, A.A.; Halley, S.; Chandler, D.; Gunson, R.; Shepherd, S.J.; Parcell, B.J. First report of an astrovirus type 5 gastroenteritis outbreak in a residential elderly care home identified by sequencing. J. Clin. Virol. 2015, 73, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Grohmann, G.S.; Glass, R.I.; Pereira, H.G.; Monroe, S.S.; Hightower, A.W.; Weber, R.; Bryan, R.T. Enteric viruses and diarrhea in HIV-infected patients. N. Engl. J. Med. 1993, 329, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Liste, M.B.; Natera, I.; Suarez, J.A.; Pujol, F.H.; Liprandi, F.; Ludert, J.E. Enteric virus infections and diarrhea in healthy and human immunodeficiency virus-infected children. J. Clin. Microbiol. 2000, 38, 2873–2877. [Google Scholar] [PubMed]
- Rossit, A.R.; de Almeida, M.T.; Nogueira, C.A.; da Costa Oliveira, J.G.; Barbosa, D.M.; Moscardini, A.C.; Mascarenhas, J.D.; Gabbay, Y.B.; Marques, F.R.; Cardoso, L.V.; et al. Bacterial, yeast, parasitic, and viral enteropathogens in HIV-infected children from Sao Paulo state, Southeastern Brazil. Diagn. Microbiol. Infect. Dis. 2007, 57, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Osborne, C.M.; Montano, A.C.; Robinson, C.C.; Schultz-Cherry, S.; Dominguez, S.R. Viral gastroenteritis in children in Colorado 2006–2009. J. Med. Virol. 2015, 87, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, S.R.; Allred, A.F.; Tarr, P.I.; Klein, E.J.; Kirkwood, C.D.; Wang, D. Metagenomic analysis of human diarrhea: Viral detection and discovery. PLoS Pathog. 2008, 4, e1000011. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, S.R.; Kirkwood, C.D.; Wang, D. Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. Virol. J. 2008, 5, 117. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, S.R.; Li, Y.; Ruone, S.; Conrardy, C.; Gregoricus, N.; Toney, D.; Virgin, H.W.; Anderson, L.J.; Vinje, J.; Wang, D.; et al. Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J. Virol. 2009, 83, 10836–10839. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, S.R.; Holtz, L.R.; Jiang, Y.; Rajendran, P.; Franz, C.J.; Zhao, G.; Kang, G.; Wang, D. Human stool contains a previously unrecognized diversity of novel astroviruses. Virol. J. 2009, 6, 161. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, S.R.; Le, B.M.; Holtz, L.R.; Storch, G.A.; Wang, D. Detection of newly described astrovirus MLB1 in stool samples from children. Emerg. Infect. Dis. 2009, 15, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Li, L.; Victoria, J.; Oderinde, B.; Mason, C.; Pandey, P.; Zaidi, S.Z.; Delwart, E. Multiple novel astrovirus species in human stool. J. Gen. Virol. 2009, 90, 2965–2972. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.L.; Cordey, S.; Brito, F.; Kaiser, L. Novel human astroviruses: Novel human diseases? J. Clin. Virol. 2016, 82, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Pinto, P.; Tummolo, F.; De Grazia, S.; Giammanco, G.M.; Medici, M.C.; Ganesh, B.; L'Homme, Y.; Farkas, T.; Jakab, F.; et al. Analysis of the ORF2 of human astroviruses reveals lineage diversification, recombination and rearrangement and provides the basis for a novel sub-classification system. Arch. Virol. 2014, 159, 3185–3196. [Google Scholar] [CrossRef] [PubMed]
- Medici, M.C.; Tummolo, F.; Martella, V.; Banyai, K.; Bonerba, E.; Chezzi, C.; Arcangeletti, M.C.; De Conto, F.; Calderaro, A. Genetic heterogeneity and recombination in type-3 human astroviruses. Infect. Genet. Evol. 2015, 32, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Kroneman, A.; Vega, E.; Vennema, H.; Vinje, J.; White, P.A.; Hansman, G.; Green, K.; Martella, V.; Katayama, K.; Koopmans, M. Proposal for a unified norovirus nomenclature and genotyping. Arch. Virol. 2013, 158, 2059–2068. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Holtz, L.R.; Bauer, I.; Franz, C.J.; Zhao, G.; Bodhidatta, L.; Shrestha, S.K.; Kang, G.; Wang, D. Comparison of novel MLB-clade, VA-clade and classic human astroviruses highlights constrained evolution of the classic human astrovirus nonstructural genes. Virology 2013, 436, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.T.; Bauer, I.K.; Antonio, M.; Adeyemi, M.; Saha, D.; Oundo, J.O.; Ochieng, J.B.; Omore, R.; Stine, O.C.; Wang, D.; et al. Prevalence of classic, MLB-clade and VA-clade astroviruses in Kenya and the Gambia. Virol. J. 2015, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.G.; Nordgren, J.; Ouermi, D.; Simpore, J.; Nitiema, L.W.; Deng, X.; Delwart, E. New astrovirus in human feces from Burkina Faso. J. Clin. Virol. 2014, 60, 161–164. [Google Scholar] [CrossRef] [PubMed]
- De Benedictis, P.; Schultz-Cherry, S.; Burnham, A.; Cattoli, G. Astrovirus infections in humans and animals—Molecular biology, genetic diversity, and interspecies transmissions. Infect. Genet. Evol. 2011, 11, 1529–1544. [Google Scholar] [CrossRef] [PubMed]
- Ulloa, J.C.; Gutierrez, M.F. Genomic analysis of two ORF2 segments of new porcine astrovirus isolates and their close relationship with human astroviruses. Can. J. Microbiol. 2010, 56, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lau, S.K.; Teng, J.L.; Tsang, A.K.; Joseph, S.; Xie, J.; Jose, S.; Fan, R.Y.; Wernery, U.; Yuen, K.Y. A novel astrovirus from dromedaries in the Middle East. J. Gen. Virol. 2015, 96, 2697–2707. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, B.; Yue, H.; Wang, Y.; Zhou, F.; Zhang, Q.; Tang, C. A novel astrovirus species in the gut of yaks with diarrhoea in the Qinghai-Tibetan Plateau, 2013. J. Gen. Virol. 2015, 96, 3672–3680. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Chmura, A.A.; Li, J.; Zhu, G.; Desmond, J.S.; Zhang, Y.; Zhang, W.; Epstein, J.H.; Daszak, P.; Shi, Z. Detection of diverse novel astroviruses from small mammals in China. J. Gen. Virol. 2014, 95, 2442–2449. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Battisti, C.; Salviato, A.; Jonassen, C.; Toffan, A.; Capua, I.; Cattoli, G. Genetic characterization of astroviruses detected in guinea fowl (Numida meleagris) reveals a distinct genotype and suggests cross-species transmission between turkey and guinea fowl. Arch. Virol. 2012, 157, 1329–1337. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, E.A.; Small, C.T.; Freiden, P.; Feeroz, M.M.; Matsen, F.A.t.; San, S.; Hasan, M.K.; Wang, D.; Jones-Engel, L.; Schultz-Cherry, S. Non-Human primates harbor diverse mammalian and avian astroviruses including those associated with human infections. PLoS Pathog. 2015, 11, e1005225. [Google Scholar] [CrossRef] [PubMed]
- Meliopoulos, V.A.; Kayali, G.; Burnham, A.; Oshansky, C.M.; Thomas, P.G.; Gray, G.C.; Beck, M.A.; Schultz-Cherry, S. Detection of antibodies against turkey astrovirus in humans. PLoS ONE 2014, 9, e96934. [Google Scholar] [CrossRef] [PubMed]
- Oude Munnink, B.B.; Cotten, M.; Canuti, M.; Deijs, M.; Jebbink, M.F.; van Hemert, F.J.; Phan, M.V.T.; Bakker, M.; Jazaeri Farsani, S.M.; Kellam, P.; et al. A novel astrovirus-like RNA virus detected in human stool. Virus Evol. 2016, 2, vew005. [Google Scholar] [CrossRef] [PubMed]
- Meliopoulos, V.A.; Marvin, S.A.; Freiden, P.; Moser, L.A.; Nighot, P.; Ali, R.; Blikslager, A.; Reddivari, M.; Heath, R.J.; Koci, M.D.; et al. Oral administration of astrovirus capsid protein is sufficient to induce acute diarrhea in vivo. mBio 2016, 7, e01494-16. [Google Scholar] [CrossRef] [PubMed]
- Moser, L.A.; Carter, M.; Schultz-Cherry, S. Astrovirus increases epithelial barrier permeability independently of viral replication. J. Virol. 2007, 81, 11937–11945. [Google Scholar] [CrossRef] [PubMed]
- Bagci, S.; Eis-Hubinger, A.M.; Yassin, A.F.; Simon, A.; Bartmann, P.; Franz, A.R.; Mueller, A. Clinical characteristics of viral intestinal infection in preterm and term neonates. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Chappe, C.; Minjolle, S.; Dabadie, A.; Morel, L.; Colimon, R.; Pladys, P. Astrovirus and digestive disorders in neonatal units. Acta Paediatr. 2012, 101, e208–e212. [Google Scholar] [CrossRef] [PubMed]
- Holtz, L.R.; Bauer, I.K.; Rajendran, P.; Kang, G.; Wang, D. Astrovirus MLB1 is not associated with diarrhea in a cohort of Indian children. PLoS ONE. 2011, 6, e28647. [Google Scholar] [CrossRef] [PubMed]
- Kapusinszky, B.; Minor, P.; Delwart, E. Nearly constant shedding of diverse enteric viruses by two healthy infants. J. Clin. Microbiol. 2012, 50, 3427–3434. [Google Scholar] [CrossRef] [PubMed]
- Resque, H.R.; Munford, V.; Castilho, J.G.; Schmich, H.; Caruzo, T.A.; Racz, M.L. Molecular characterization of astrovirus in stool samples from children in Sao Paulo, Brazil. Mem. Inst. Oswaldo Cruz 2007, 102, 969–974. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, P.; Payne, D.C.; Szilagyi, P.G.; Edwards, K.M.; Staat, M.A.; Shirley, S.H.; Wikswo, M.; Nix, W.A.; Lu, X.; Parashar, U.D.; et al. Etiology of viral gastroenteritis in children <5 years of age in the United States, 2008–2009. J. Infect. Dis. 2013, 208, 790–800. [Google Scholar] [PubMed]
- Moser, L.A.; Schultz-Cherry, S. Pathogenesis of astrovirus infection. Viral Immunol. 2005, 18, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Wunderli, W.; Meerbach, A.; Gungor, T.; Berger, C.; Greiner, O.; Caduff, R.; Trkola, A.; Bossart, W.; Gerlach, D.; Schibler, M.; et al. Astrovirus infection in hospitalized infants with severe combined immunodeficiency after allogeneic hematopoietic stem cell transplantation. PLoS ONE 2011, 6, e27483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mor, S.K.; Abin, M.; Costa, G.; Durrani, A.; Jindal, N.; Goyal, S.M.; Patnayak, D.P. The role of type-2 turkey astrovirus in poult enteritis syndrome. Poult. Sci. 2011, 90, 2747–2752. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, B.O.; Backhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 2016, 22, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Pativada, M.; Nataraju, S.M.; Ganesh, B.; Rajendran, K.; Ramamurthy, T.; Ganguly, S.; Bhattacharya, M.K.; Ghosh, M.; Kobayashi, N.; Krishnan, T. Emerging trends in the epidemiology of human astrovirus infection among infants, children and adults hospitalized with acute watery diarrhea in Kolkata, India. Infect. Genet. Evol. 2012, 12, 1685–1693. [Google Scholar] [CrossRef]
- Pfeiffer, J.K.; Virgin, H.W. Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science 2016, 351, 6270. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, M.A.; Saif, Y.M.; Heggen-Peay, C.L.; Edens, F.W.; Havenstein, G.B. Induction of functional defects in macrophages by a poult enteritis and mortality syndrome-associated turkey astrovirus. Avian Dis. 2001, 45, 853–861. [Google Scholar] [CrossRef] [PubMed]
- Quan, P.L.; Wagner, T.A.; Briese, T.; Torgerson, T.R.; Hornig, M.; Tashmukhamedova, A.; Firth, C.; Palacios, G.; Baisre-De-Leon, A.; Paddock, C.D.; et al. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg. Infect. Dis. 2010, 16, 918–925. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R.; Morfopoulou, S.; Hubb, J.; Emmett, W.A.; Ip, W.; Shah, D.; Brooks, T.; Paine, S.M.; Anderson, G.; Virasami, A.; et al. Astrovirus VA1/HMO-C: An increasingly recognized neurotropic pathogen in immunocompromised patients. Clin. Infect. Dis. 2015, 60, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Naccache, S.N.; Peggs, K.S.; Mattes, F.M.; Phadke, R.; Garson, J.A.; Grant, P.; Samayoa, E.; Federman, S.; Miller, S.; Lunn, M.P.; et al. Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-generation sequencing. Clin. Infect. Dis. 2015, 60, 919–923. [Google Scholar] [CrossRef] [PubMed]
- Fremond, M.L.; Perot, P.; Muth, E.; Cros, G.; Dumarest, M.; Mahlaoui, N.; Seilhean, D.; Desguerre, I.; Hebert, C.; Corre-Catelin, N.; et al. Next-Generation sequencing for diagnosis and tailored therapy: A case report of astrovirus-associated progressive encephalitis. J. Pediatric. Infect. Dis. Soc. 2015, 4, e53–e57. [Google Scholar] [CrossRef] [PubMed]
- Lum, S.H.; Turner, A.; Guiver, M.; Bonney, D.; Martland, T.; Davies, E.; Newbould, M.; Brown, J.; Morfopoulou, S.; Breuer, J.; et al. An emerging opportunistic infection: Fatal astrovirus (VA1/HMO-C) encephalitis in a pediatric stem cell transplant recipient. Transpl. Infect. Dis. 2016. [Google Scholar] [CrossRef] [PubMed]
- Cordey, S.; Vu, D.-L.; Schibler, M.; L'Huillier, A.G.; Brito, F.; Docquier, M.; Posfay-Barbe, K.M.; Petty, T.J.; Turin, L.; Zdobnov, E.M.; et al. Astrovirus MLB2: A new gastro-enteric virus associated with meningitis and disseminated infection. Emerg. Infect. Dis. 2016, 22, 846–853. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Kuroda, M.; Kasai, M.; Matsui, H.; Fukuyama, T.; Katano, H.; Tanaka-Taya, K. Acute encephalopathy in an immunocompromised boy with astrovirus-MLB1 infection detected by next generation sequencing. J. Clin. Virol. 2016, 78, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Wuthrich, D.; Boujon, C.L.; Truchet, L.; Selimovic-Hamza, S.; Oevermann, A.; Bouzalas, I.G.; Bruggmann, R.; Seuberlich, T. Exploring the virome of cattle with non-suppurative encephalitis of unknown etiology by metagenomics. Virology 2016, 493, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Blomstrom, A.L.; Widen, F.; Hammer, A.S.; Belak, S.; Berg, M. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J. Clin. Microbiol. 2010, 48, 4392–4396. [Google Scholar] [CrossRef] [PubMed]
- Blomstrom, A.L.; Ley, C.; Jacobson, M. Astrovirus as a possible cause of congenital tremor type AII in piglets? Acta Vet. Scand. 2014, 56, 82. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.J.; David, T.J.; Chrystie, I.L.; Totterdell, B. Chronic enteric virus infection in two T-cell immunodeficient children. J. Med. Virol. 1988, 24, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, C.C.; Loh, J.; Zhao, G.; Stappenbeck, T.S.; Wang, D.; Huang, H.V.; Virgin, H.W.; Thackray, L.B. Adaptive immunity restricts replication of novel murine astroviruses. J. Virol. 2012, 86, 12262–12270. [Google Scholar] [CrossRef] [PubMed]
- Koopmans, M.P.; Bijen, M.H.; Monroe, S.S.; Vinje, J. Age-Stratified seroprevalence of neutralizing antibodies to astrovirus types 1 to 7 in humans in the Netherlands. Clin. Diagn. Lab. Immunol. 1998, 5, 33–37. [Google Scholar] [PubMed]
- Guix, S.; Pérez-Bosque, A.; Miró, L.; Moretó, M.; Bosch, A.; Pintó, R.M. Type I interferon response is delayed in human astrovirus infections. PLoS ONE 2015, 10, e0123087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marvin, S.A.; Huerta, C.T.; Sharp, B.; Freiden, P.; Cline, T.D.; Schultz-Cherry, S. Type I interferon response limits astrovirus replication and protects against increased barrier permeability in vitro and in vivo. J. Virol. 2015, 90, 1988–1996. [Google Scholar] [CrossRef] [PubMed]
- Koci, M.D.; Kelley, L.A.; Larsen, D.; Schultz-Cherry, S. Astrovirus-Induced synthesis of nitric oxide contributes to virus control during infection. J. Virol. 2004, 78, 1564–1574. [Google Scholar] [CrossRef] [PubMed]
- Holtz, L.R.; Wylie, K.M.; Sodergren, E.; Jiang, Y.; Franz, C.J.; Weinstock, G.M.; Storch, G.A.; Wang, D. Astrovirus MLB2 viremia in febrile child. Emerg. Infect. Dis. 2011, 17, 2050–2052. [Google Scholar] [CrossRef] [PubMed]
- Wylie, K.M.; Mihindukulasuriya, K.A.; Sodergren, E.; Weinstock, G.M.; Storch, G.A. Sequence analysis of the human virome in febrile and afebrile children. PLoS ONE 2012, 7, e27735. [Google Scholar] [CrossRef] [PubMed]
- Cordey, S.; Brito, F.; Vu, D.L.; Turin, L.; Kilowoko, M.; Kyungu, E.; Genton, B.; Zdobnov, E.M.; D'Acremont, V.; Kaiser, L. Astrovirus VA1 identified by next-generation sequencing in a nasopharyngeal specimen of a febrile tanzanian child with acute respiratory disease of unknown etiology. Emerg. Microbes Infect. 2016, 5, e99. [Google Scholar] [CrossRef] [PubMed]
- Meliopoulos, V.; Schultz-Cherry, S. Astrovirus pathogenesis. In Astrovirus Research: Essential Ideas, Everyday Impacts, Future Directions; Schultz-Cherry, S., Ed.; Springer: New York, NY, USA, 2013; pp. 65–77. [Google Scholar]
- Pantin-Jackwood, M.; Todd, D.; Koci, M.D. Avian astroviruses. In Astrovirus Research: Essential Ideas, Everyday Impacts, Future Directions; Schultz-Cherry, S., Ed.; Springer: New York, NY, USA, 2013; pp. 151–180. [Google Scholar]
- Wang, Y.; Li, Y.; Jin, Y.; Li, D.D.; Li, X.; Duan, Z.J. Recently identified novel human astroviruses in children with diarrhea, China. Emerg. Infect. Dis. 2013, 19, 1333–1335. [Google Scholar] [CrossRef] [PubMed]
- Xavier, M.d.P.T.P.; Carvalho Costa, F.A.; Rocha, M.S.; Andrade, J.d.S.R.d.; Diniz, F.K.B.; Andrade, T.R.d.; Miagostovich, M.P.; Leite, J.P.G.; Volotão, E.d.M. Surveillance of human astrovirus infection in Brazil: The first report of MLB1 astrovirus. PLoS ONE 2015, 10, e0135687. [Google Scholar] [CrossRef] [PubMed]
- Galdiero, E.; Marinelli, A.; Pisciotta, M.G.; Pagliara, I.; Di Monteforte, E.S.; Liguori, G. Reverse transcriptase-PCR for the detection of astrovirus in children with nosocomial acute diarrhoea in Naples, Italy. Méd. Mal. Infect. 2005, 35, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Nigel, A.C.; Booth, J.A.; Claire, E.; Sharon, J.L.; Will, S.; Nick, K.; Osamu, N.; Toyoko, N.; Hart, C.A.; Martyn, R. Healthcare-Associated viral gastroenteritis among children in a large pediatric hospital, United Kingdom. Emerg. Infect. Dis. J 2010, 16, 55. [Google Scholar]
- Nguyen, T.A.; Yagyu, F.; Okame, M.; Phan, T.G.; Trinh, Q.D.; Yan, H.; Hoang, K.T.; Cao, A.T.; Le Hoang, P.; Okitsu, S.; et al. Diversity of viruses associated with acute gastroenteritis in children hospitalized with diarrhea in Ho Chi Minh city, Vietnam. J. Med. Virol. 2007, 79, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Rimoldi, S.G.; Stefani, F.; Pagani, C.; Chenal, L.L.; Zanchetta, N.; Di Bartolo, I.; Lombardi, A.; Ruggeri, F.M.; Di lillo, D.; Zuccotti, G.V.; et al. Epidemiological and clinical characteristics of pediatric gastroenteritis associated with new viral agents. Arch. Virol. 2011, 156, 1583. [Google Scholar] [CrossRef] [PubMed]
- Al-Thani, A.; Baris, M.; Al-Lawati, N.; Al-Dhahry, S. Characterising the aetiology of severe acute gastroenteritis among patients visiting a hospital in Qatar using real-time polymerase chain reaction. BMC Infect. Dis. 2013, 13, 329. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, G.G.; Liprandi, F.; Ludert, J.E. Molecular epidemiology of enteric viruses in children with sporadic gastroenteritis in Valencia, Venezuela. J. Med. Virol. 2011, 83, 1972–1982. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, Y.-H.; Peng, J.-S.; Zhou, X.; Tang, L.; Kobayashi, N.; Hu, Q.; Zhou, D.-J.; Huang, H.-J.; Liu, M.-Q. Genetic characterization of human astrovirus infection in Wuhan, People’s Republic of China, 2007–2008. Can. J. Microbiol. 2011, 57, 964–968. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Ma, H.; Jin, M.; Wang, X.; Wang, J.; Xu, L.; Lin, S.; Shen, Z.; Chen, Z.; Qiu, Z.; et al. Etiology and epidemiology of viral diarrhea in children under the age of five hospitalized in Tianjin, China. Arch. Virol. 2012, 157, 881–887. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Jia, R.; Zhong, H.; Xu, M.; Su, L.; Cao, L.; Dong, Z.; Dong, N.; Xu, J. Molecular characterization and multiple infections of rotavirus, norovirus, sapovirus, astrovirus and adenovirus in outpatients with sporadic gastroenteritis in Shanghai, China, 2010–2011. Arch. Virol. 2015, 160, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.-c.; Xu, Q.-h.; Wu, X.-b.; Hu, G.-f.; Tang, Y.-l.; Li, J.-d.; Chen, Q.; Nie, J. Development of real-time and nested RT-PCR to detect astrovirus and one-year survey of astrovirus in Jiangmen city, China. Arch. Virol. 2010, 155, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Verma, H.; Chitambar, S.D.; Gopalkrishna, V. Astrovirus associated acute gastroenteritis in western India: Predominance of dual serotype strains. Infect. Genet. Evol. 2010, 10, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Thongprachum, A.; Takanashi, S.; Kalesaran, A.F.C.; Okitsu, S.; Mizuguchi, M.; Hayakawa, S.; Ushijima, H. Four-Year study of viruses that cause diarrhea in japanese pediatric outpatients. J. Med. Virol. 2015, 87, 1141–1148. [Google Scholar] [CrossRef] [PubMed]
- Yoneda, M.; Nakano, M.; Sugimoto, D.; Inada, M.; Fujitani, M.; Kitahori, Y. Epidemiological characteristics of sapovirus and human astrovirus detected among children in the Nara Prefecture, Japan, during 2009/2010–2014/2015 seasons. Jpn. J. Infect. Dis. 2016, 70, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, N.; Kobayashi, S.; Minagawa, H.; Matsushita, T.; Sugiura, W.; Iwatani, Y. Molecular epidemiology of enteric viruses in patients with acute gastroenteritis in Aichi Prefecture, Japan, 2008/09−2013/14. J. Med. Virol. 2016, 88, 1180–1186. [Google Scholar] [CrossRef] [PubMed]
- Khamrin, P.; Thongprachum, A.; Okitsu, S.; Hayakawa, S.; Maneekarn, N.; Ushijima, H. Multiple astrovirus MLB1, MLB2, VA2 clades, and classic human astrovirus in children with acute gastroenteritis in Japan. J. Med. Virol. 2016, 88, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Wu, F.T.; Huang, Y.C.; Chang, W.C.; Wu, H.S.; Wu, C.Y.; Lin, J.S.; Huang, F.C.; Hsiung, C.A. Clinical and epidemiologic features of severe viral gastroenteritis in children: A 3-year surveillance, multicentered study in Taiwan with partial rotavirus immunization. Med. (Baltim.) 2015, 94, e1372. [Google Scholar] [CrossRef] [PubMed]
- Malasao, R.; Khamrin, P.; Chaimongkol, N.; Ushijima, H.; Maneekarn, N. Diversity of human astrovirus genotypes circulating in children with acute gastroenteritis in Thailand during 2000–2011. J. Med. Virol. 2012, 84, 1751–1756. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.A.; Hoang, L.; Pham le, D.; Hoang, K.T.; Mizuguchi, M.; Okitsu, S.; Ushijima, H. Identification of human astrovirus infections among children with acute gastroenteritis in the Southern part of Vietnam during 2005–2006. J. Med. Virol. 2008, 80, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Ouédraogo, N.; Kaplon, J.; Bonkoungou, I.J.O.; Traoré, A.S.; Pothier, P.; Barro, N.; Ambert-Balay, K. Prevalence and genetic diversity of enteric viruses in children with diarrhea in Ouagadougou, Burkina Faso. PLoS ONE 2016, 11, e0153652. [Google Scholar] [CrossRef] [PubMed]
- Lekana-Douki, S.E.; Kombila-Koumavor, C.; Nkoghe, D.; Drosten, C.; Drexler, J.F.; Leroy, E.M. Molecular epidemiology of enteric viruses and genotyping of rotavirus a, adenovirus and astrovirus among children under 5 years old in Gabon. Int. J. Infect. Dis. 2015, 34, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Reither, K.; Ignatius, R.; Weitzel, T.; Seidu-Korkor, A.; Anyidoho, L.; Saad, E.; Djie-Maletz, A.; Ziniel, P.; Amoo-Sakyi, F.; Danikuu, F.; et al. Acute childhood diarrhoea in northern Ghana: Epidemiological, clinical and microbiological characteristics. BMC Infect. Dis. 2007, 7, 104. [Google Scholar] [CrossRef] [PubMed]
- Mladenova, Z.; Steyer, A.; Steyer, A.F.; Ganesh, B.; Petrov, P.; Tchervenjakova, T.; Iturriza-Gomara, M. Aetiology of acute paediatric gastroenteritis in Bulgaria during summer months: Prevalence of viral infections. J. Med. Microbiol. 2015, 64, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Simonen-Tikka, M.-L.; Klemola, P.; Suomenrinne, S.; Kaijalainen, S.; Söderström, D.; Savolainen-Kopra, C.; Näntö-Salonen, K.; Ilonen, J.; Simell, T.; Simell, O.; et al. Virus infections among young children—The first year of the indis study. J. Med. Virol. 2013, 85, 1678–1684. [Google Scholar] [CrossRef] [PubMed]
- Medici, M.C.; Tummolo, F.; Albonetti, V.; Abelli, L.A.; Chezzi, C.; Calderaro, A. Molecular detection and epidemiology of astrovirus, bocavirus, and sapovirus in italian children admitted to hospital with acute gastroenteritis, 2008–2009. J. Med. Virol. 2012, 84, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, P.; Samoilovich, E.; Yermalovich, M.; Chernyshova, L.; Gheorghita, S.; Cojocaru, R.; Shugayev, N.; Sahakyan, G.; Lashkarashvili, M.; Chubinidze, M.; et al. Viral gastroenteritis in rotavirus negative hospitalized children <5 years of age from the independent states of the former Soviet Union. Infect. Genet. Evol. 2014, 28, 283–288. [Google Scholar] [PubMed]
- Gabbay, Y.B.; Luz, C.R.; Costa, I.V.; Cavalcante-Pepino, E.L.; Sousa, M.S.; Oliveira, K.K.; Wanzeller, A.L.; Mascarenhas, J.D.; Leite, J.P.; Linhares, A.C. Prevalence and genetic diversity of astroviruses in children with and without diarrhea in Sao Luis, Maranhao, Brazil. Mem. Inst. Oswaldo Cruz 2005, 100, 709–714. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.A.; Borges, A.M.; da Costa, P.S.; Teixeira, J.M.; Giugliano, L.G.; Leite, J.P.; Cardoso, D. Astrovirus infection in children living in the central west region of Brazil. Mem. Inst. Oswaldo Cruz 2007, 102, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-J.; Liu, W.; Liu, Y.-X.; Xiao, H.-J.; Jia, N.; Liu, G.; Tong, Y.-G.; Cao, W.-C. Identification of norovirus as the top enteric viruses detected in adult cases with acute gastroenteritis. Am. J. Trop. Med. Hyg. 2010, 82, 717–722. [Google Scholar] [PubMed]
- Arena, C.; Amoros, J.P.; Vaillant, V.; Ambert-Balay, K.; Chikhi-Brachet, R.; Jourdan-Da Silva, N.; Varesi, L.; Arrighi, J.; Souty, C.; Blanchon, T.; et al. Acute diarrhea in adults consulting a general practitioner in France during winter: Incidence, clinical characteristics, management and risk factors. BMC Infect. Dis. 2014, 14, 574. [Google Scholar] [CrossRef] [PubMed]
- Podkolzin, A.T.; Fenske, E.B.; Abramycheva, N.Y.; Shipulin, G.A.; Sagalova, O.I.; Mazepa, V.N.; Ivanova, G.N.; Semena, A.V.; Tagirova, Z.G.; Alekseeva, M.N.; et al. Hospital-Based surveillance of rotavirus and other viral agents of diarrhea in children and adults in Russia, 2005–2007. J. Infect. Dis. 2009, 200 (Suppl. 1), S228–S233. [Google Scholar] [CrossRef] [PubMed]
- Chau, M.L.; Hartantyo, S.H.; Yap, M.; Kang, J.S.; Aung, K.T.; Gutierrez, R.A.; Ng, L.C.; Tam, C.C.; Barkham, T. Diarrheagenic pathogens in adults attending a hospital in Singapore. BMC Infect. Dis. 2016, 16, 32. [Google Scholar] [CrossRef] [PubMed]
- Gabbay, Y.B.; Leite, J.P.; Oliveira, D.S.; Nakamura, L.S.; Nunes, M.R.; Mascarenhas, J.D.; Heinemann, M.B.; Linhares, A.C. Molecular epidemiology of astrovirus type 1 in Belem, Brazil, as an agent of infantile gastroenteritis, over a period of 18 years (1982–2000): Identification of two possible new lineages. Virus Res. 2007, 129, 166–174. [Google Scholar] [CrossRef] [PubMed]
- De Grazia, S.; Bonura, F.; Bányai, K.; Gellért, Á.; Marineo, S.; Martella, V.; Giammanco, G.M. Temporal variation in the distribution of type-1 human astrovirus lineages in a settled population over 14 years. Arch. Virol. 2016, 161, 1633–1637. [Google Scholar] [CrossRef] [PubMed]
- Grant, L.; Vinje, J.; Parashar, U.; Watt, J.; Reid, R.; Weatherholtz, R.; Santosham, M.; Gentsch, J.; O'Brien, K. Epidemiologic and clinical features of other enteric viruses associated with acute gastroenteritis in american indian infants. J. Pediatr. 2012, 161, 110–115; e111. [Google Scholar] [CrossRef] [PubMed]
- Platts-Mills, J.A.; Gratz, J.; Mduma, E.; Svensen, E.; Amour, C.; Liu, J.; Maro, A.; Saidi, Q.; Swai, N.; Kumburu, H.; et al. Association between stool enteropathogen quantity and disease in tanzanian children using Taqman array cards: A nested case-control study. Am. J. Trop. Med. Hyg. 2014, 90, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Lopman, B.A.; Steele, D.; Kirkwood, C.D.; Parashar, U.D. The vast and varied global burden of norovirus: Prospects for prevention and control. PLoS Med. 2016, 13, e1001999. [Google Scholar] [CrossRef]
- Svraka, S.; Duizer, E.; Vennema, H.; de Bruin, E.; van der Veer, B.; Dorresteijn, B.; Koopmans, M. Etiological role of viruses in outbreaks of acute gastroenteritis in the Netherlands from 1994 through 2005. J. Clin. Microbiol. 2007, 45, 1389–1394. [Google Scholar] [CrossRef] [PubMed]
- Lyman, W.H.; Walsh, J.F.; Kotch, J.B.; Weber, D.J.; Gunn, E.; Vinje, J. Prospective study of etiologic agents of acute gastroenteritis outbreaks in child care centers. J. Pediatr. 2009, 154, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.A.; Bruggink, L.D.; Sturge, K.; Subasinghe, N.; Tan, A.; Hogg, G.G. Molecular features of astrovirus associated with a gastroenteritis outbreak in an aged-care centre. Eur. J. Clin. Microbiol. Infect. Dis. 2007, 26, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Oishi, I.; Yamazaki, K.; Kimoto, T.; Minekawa, Y.; Utagawa, E.; Yamazaki, S.; Inouye, S.; Grohmann, G.S.; Monroe, S.S.; Stine, S.E.; et al. A large outbreak of acute gastroenteritis associated with astrovirus among students and teachers in Osaka, Japan. J. Infect. Dis. 1994, 170, 439–443. [Google Scholar] [CrossRef] [PubMed]
- van der Doef, H.P.J.; Bathoorn, E.; van der Linden, M.P.M.; Wolfs, T.F.W.; Minderhoud, A.L.C.; Bierings, M.B.; Wensing, A.M.J.; Lindemans, C.A. Astrovirus outbreak at a pediatric hematology and hematopoietic stem cell transplant unit despite strict hygiene rules. Bone Marrow. Transpl. 2016, 51, 747–750. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-y.; Liu, N.; Guo, W.-d.; Yu, Q.; Wang, W.-r.; Song, Z.-Z.; Yan, H.; Luo, Y.; Lu, A.-t.; Li, H.-Y.; et al. Outbreak of neonatal gastroenteritis associated with astrovirus serotype 1 at a hospital in Inner Mongolia, China. J. Clin. Microbiol. 2010, 48, 4306–4309. [Google Scholar] [CrossRef] [PubMed]
- Sirinavin, S.; Techasaensiri, C.; Okascharoen, C.; Nuntnarumit, P.; Tonsuttakul, S.; Pongsuwan, Y. Neonatal astrovirus gastroenteritis during an inborn nursery outbreak. J. Hosp. Infect. 2006, 64, 196–197. [Google Scholar] [CrossRef] [PubMed]
- Hwang, B.J.S.; Jeong, H.J.; Chung, G.T.; Kang, Y.; Yang, S.J.; Seo, N.Y.; Shin, T.H.; Yoo, C.; Lee, D.Y. Outbreak of astrovirus in adults with acute gastroenteritis in Korea. J. Gastrointest. Dig. Syst. 2015, S13. [Google Scholar] [CrossRef]
- Burbelo, P.D.; Ching, K.H.; Esper, F.; Iadarola, M.J.; Delwart, E.; Lipkin, W.I.; Kapoor, A. Serological studies confirm the novel astrovirus HMOAstV-C as a highly prevalent human infectious agent. PLoS ONE 2011, 6, e22576. [Google Scholar] [CrossRef] [PubMed]
- Holtz, L.R.; Bauer, I.K.; Jiang, H.; Belshe, R.; Freiden, P.; Schultz-Cherry, S.L.; Wang, D. Seroepidemiology of astrovirus MLB1. Clin. Vaccine Immunol. 2014, 21, 908–911. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.K.W.; Chin, A.W.H.; Smith, G.J.; Chan, K.-H.; Guan, Y.; Peiris, J.S.M.; Poon, L.L.M. Detection of novel astroviruses in urban brown rats and previously known astroviruses in humans. J. Gen. Virol. 2010, 91, 2457–2462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, S.F.; Sebeny, P.J.; Klena, J.D.; Pimentel, G.; Mansour, A.; Naguib, A.M.; Bruton, J.; Young, S.Y.; Holtz, L.R.; Wang, D. Novel astroviruses in children, Egypt. Emerg. Infect. Dis. 2011, 17, 2391–2393. [Google Scholar] [CrossRef] [PubMed]
- Mitui, M.T.; Bozdayi, G.; Matsumoto, T.; Dalgic, B.; Nishizono, A.; Ahmed, K. Complete genome sequence of an MLB2 astrovirus from a turkish child with diarrhea. Genome Announc. 2013, 1. [Google Scholar] [CrossRef] [PubMed]
- Midthun, K.; Greenberg, H.B.; Kurtz, J.B.; Gary, G.W.; Lin, F.Y.; Kapikian, A.Z. Characterization and seroepidemiology of a type 5 astrovirus associated with an outbreak of gastroenteritis in Marin County, California. J. Clin. Microbiol. 1993, 31, 955–962. [Google Scholar] [PubMed]
- Kurtz, J.B.; Lee, T.W.; Craig, J.W.; Reed, S.E. Astrovirus infection in volunteers. J. Med. Virol. 1979, 3, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Abad, F.X.; Pinto, R.M.; Villena, C.; Gajardo, R.; Bosch, A. Astrovirus survival in drinking water. Appl. Environ. Microbiol. 1997, 63, 3119–3122. [Google Scholar] [PubMed]
- Bosch, A.; Pinto, R.M.; Villena, C.; Abad, F.X. Persistence of human astrovirus in fresh and marine water. Water Sci. Technol. 1997, 35, 243–247. [Google Scholar] [CrossRef]
- Pintó, R.M.; Villena, C.; Le Guyader, F.; Guix, S.; Caballero, S.; Pommepuy, M.; Bosch, A. Astrovirus detection in wastewater samples. Water Sci. Technol. 2001, 43, 73–76. [Google Scholar] [PubMed]
- Le Cann, P.; Ranarijaona, S.; Monpoeho, S.; Le Guyader, F.; Ferre, V. Quantification of human astroviruses in sewage using real-time RT-PCR. Res. Microbiol. 2004, 155, 11–15. [Google Scholar] [CrossRef] [PubMed]
- El-Senousy, W.M.; Guix, S.; Abid, I.; Pinto, R.M.; Bosch, A. Removal of astrovirus from water and sewage treatment plants, evaluated by a competitive reverse transcription-PCR. Appl. Environ. Microbiol. 2007, 73, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Prevost, B.; Lucas, F.S.; Ambert-Balay, K.; Pothier, P.; Moulin, L.; Wurtzer, S. Deciphering the diversities of astroviruses and noroviruses in wastewater treatment plant effluents by a high-throughput sequencing method. Appl. Environ. Microbiol. 2015, 81, 7215–7222. [Google Scholar] [CrossRef] [PubMed]
- Lizasoain, A.; Tort, L.F.; Garcia, M.; Gomez, M.M.; Leite, J.P.; Miagostovich, M.P.; Cristina, J.; Colina, R.; Victoria, M. Environmental assessment reveals the presence of MLB-1 human astrovirus in Uruguay. J. Appl. Microbiol. 2015, 119, 859–867. [Google Scholar] [CrossRef] [PubMed]
- Hata, A.; Katayama, H.; Kitajima, M.; Furumai, H. Wastewater analysis indicates that genetically diverse astroviruses, including strains belonging to novel clades MLB and VA, are circulating within Japanese populations. Appl. Environ. Microbiol. 2015, 81, 4932–4939. [Google Scholar] [CrossRef] [PubMed]
- Le Guyader, F.; Haugarreau, L.; Miossec, L.; Dubois, E.; Pommepuy, M. Three-Year study to assess human enteric viruses in shellfish. Appl. Environ. Microbiol. 2000, 66, 3241–3248. [Google Scholar] [CrossRef] [PubMed]
- Nadan, S.; Walter, J.E.; Grabow, W.O.; Mitchell, D.K.; Taylor, M.B. Molecular characterization of astroviruses by reverse transcriptase PCR and sequence analysis: Comparison of clinical and environmental isolates from South Africa. Appl. Environ. Microbiol. 2003, 69, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Pintó, R.M.; Abad, F.X.; Gajardo, R.; Bosch, A. Detection of infectious astroviruses in water. Appl. Environ. Microbiol. 1996, 62, 1811–1813. [Google Scholar] [PubMed]
- World Health Organization (WHO). Guidelines for Drinking-Water Quality, Volume 1: Recommendations, 3rd ed.; WHO: Geneva, Switzerland, 2004; p. 515. [Google Scholar]
- Gofti-Laroche, L.; Gratacap-Cavallier, B.; Demanse, D.; Genoulaz, O.; Seigneurin, J.M.; Zmirou, D. Are waterborne astrovirus implicated in acute digestive morbidity (E.MI.R.A. Study)? J. Clin. Virol. 2003, 27, 74–82. [Google Scholar] [CrossRef]
- Dongdem, J.T.; Damanka, S.; Asmah, R. Molecular isolation of human norovirus and astrovirus in tap water by RT-PCR. Int. Res. J. Biochem. Bioinform. 2011, 1, 131–138. [Google Scholar]
- Steyer, A.; Torkar, K.G.; Gutiérrez-Aguirre, I.; Poljšak-Prijatelj, M. High prevalence of enteric viruses in untreated individual drinking water sources and surface water in Slovenia. Int. J. Hyg. Environ. Health 2011, 214, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Myint, S.; Manley, R.; Cubitt, D. Viruses in bathing waters. Lancet 1994, 343, 1640–1641. [Google Scholar] [CrossRef]
- Maunula, L.; Kalso, S.; von Bonsdorff, C.H.; Ponka, A. Wading pool water contaminated with both noroviruses and astroviruses as the source of a gastroenteritis outbreak. Epidemiol. Infect. 2004, 132, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Toss, M.; Griffin, D.D.; Calva, J.; Contreras, J.F.; Puerto, F.I.; Mota, F.; Guiscafre, H.; Cedillo, R.; Munoz, O.; Herrera, I.; et al. Prevalence and genetic diversity of human astroviruses in mexican children with symptomatic and asymptomatic infections. J. Clin. Microbiol. 2004, 42, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, J.E.; Taylor, D.N.; Echeverria, P.; Blacklow, N.R. Astroviruses as a cause of gastroenteritis in children. N. Engl. J. Med. 1991, 324, 1757–1760. [Google Scholar] [CrossRef] [PubMed]
- Konno, T.; Suzuki, H.; Ishida, N.; Chiba, R.; Mochizuki, K.; Tsunoda, A. Astrovirus-Associated epidemic gastroenteritis in Japan. J. Med. Virol. 1982, 9, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Todd, E.C.; Greig, J.D.; Bartleson, C.A.; Michaels, B.S. Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 3. Factors contributing to outbreaks and description of outbreak categories. J. Food Prot. 2007, 70, 2199–2217. [Google Scholar] [CrossRef] [PubMed]
- Gallimore, C.I.; Taylor, C.; Gennery, A.R.; Cant, A.J.; Galloway, A.; Lewis, D.; Gray, J.J. Use of a heminested reverse transcriptase PCR assay for detection of astrovirus in environmental swabs from an outbreak of gastroenteritis in a pediatric primary immunodeficiency unit. J. Clin. Microbiol. 2005, 43, 3890–3894. [Google Scholar] [CrossRef] [PubMed]
- Cubitt, W.D.; Mitchell, D.K.; Carter, M.J.; Willcocks, M.M.; Holzel, H. Application of electronmicroscopy, enzyme immunoassay, and RT-PCR to monitor an outbreak of astrovirus type 1 in a paediatric bone marrow transplant unit. J. Med. Virol. 1999, 57, 313–321. [Google Scholar] [CrossRef]
- Abad, F.X.; Villena, C.; Guix, S.; Caballero, S.; Pinto, R.M.; Bosch, A. Potential role of fomites in the vehicular transmission of human astroviruses. Appl. Environ. Microbiol. 2001, 67, 3904–3907. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, J.B.; Lee, T.W.; Parsons, A.J. The action of alcohols on rotavirus, astrovirus and enterovirus. J. Hosp. Infect. 1980, 1, 321–325. [Google Scholar] [CrossRef]
- Caballero, S.; Guix, S.; Ribes, E.; Bosch, A.; Pinto, R.M. Structural requirements of astrovirus virus-like particles assembled in insect cells. J. Virol. 2004, 78, 13285–13292. [Google Scholar] [CrossRef] [PubMed]
- Dalton, R.M.; Pastrana, E.P.; Sánchez-Fauquier, A. Vaccinia virus recombinant expressing an 87-kilodalton polyprotein that is sufficient to form astrovirus-like particles. J. Virol. 2003, 77, 9094–9098. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Wei, C.; Wang, L.; Cao, D.; Meng, X.J.; Jiang, X.; Tan, M. A trivalent vaccine candidate against hepatitis e virus, norovirus, and astrovirus. Vaccine 2016, 34, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Wei, C.; Wang, L.; Cao, D.; Meng, X.J.; Jiang, X.; Tan, M. Development and evaluation of two subunit vaccine candidates containing antigens of hepatitis e virus, rotavirus, and astrovirus. Sci. Rep. 2016, 6, 25735. [Google Scholar] [CrossRef] [PubMed]
- Bjorkholm, M.; Celsing, F.; Runarsson, G.; Waldenstrom, J. Successful intravenous immunoglobulin therapy for severe and persistent astrovirus gastroenteritis after fludarabine treatment in a patient with Waldenström's macroglobulinemia. Int. J. Hematol. 1995, 62, 117–120. [Google Scholar] [CrossRef]
- Superti, F.; Seganti, L.; Orsi, N.; Desideri, N.; Stein, M.L.; Tinari, A.; Marziano, M.L.; Donelli, G. In vitro effect of synthetic flavanoids on astrovirus infection. Antivir. Res. 1990, 13, 201–208. [Google Scholar] [CrossRef]
- Tellez, M.A.; Tellez, A.N.; Velez, F.; Ulloa, J.C. In vitro antiviral activity against rotavirus and astrovirus infection exerted by substances obtained from Achyrocline bogotensis (Kunth) DC. (Compositae). BMC Complement. Altern. Med. 2015, 15, 428. [Google Scholar] [CrossRef] [PubMed]
| Mamastrovirus Species | Classic | MLB | VA2-VA4 | VA1-VA3 | VA5 |
|---|---|---|---|---|---|
| Mamastrovirus 1 | Mamastrovirus 6 | Mamastrovirus 8 | Mamastrovirus 9 | Unassigned | |
| Serotypes/Clades | HAstV-1 to 8 | MLB1, MLB2 and MLB3 | VA2 (HMO-A) and VA4 | VA1 (HMO-C) and VA3 (HMO-B) | VA5 |
| ORF1a (protease and other nonstructural proteins) | |||||
| Classic | 100 | – | – | – | – |
| MLB | 32.8 | 100 | – | – | – |
| VA2-VA4 | 24.1 | 29.1 | 100 | – | – |
| VA1-VA3 | 24.2 | 28.9 | 67.4 | 100 | – |
| VA5 | 23.9 | 28.2 | 61.5 | 59.6 | 100 |
| ORF1b (RNA dependent RNA polymerase) | |||||
| Classic | 100 | – | – | – | – |
| MLB | 54.5 | 100 | – | – | – |
| VA2-VA4 | 51.8 | 49.4 | 100 | – | – |
| VA1-VA3 | 53.0 | 49.3 | 73.7 | 100 | – |
| VA5 | 50.2 | 50.7 | 74.0 | 71.5 | 100 |
| ORF2 (capsid proteins) | |||||
| Classic | 100 | – | – | – | – |
| MLB | 27.5 | 100 | – | – | – |
| VA2-VA4 | 24.0 | 21.9 | 100 | – | – |
| VA1-VA3 | 23.0 | 22.1 | 51.9 | 100 | – |
| VA5 | 23.8 | 20.6 | 58.9 | 53.1 | 100 |
| Type of Novel HAstV | Year | Country | Age of Patient | Underlying Condition | Type of CNS Infection/Presentation | Treatment | IS/Other | Outcome | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Mamastrovirus 1 | |||||||||
| HAstV-4 | 2008 | Switzerland | 3 months | HSCT for severe combined immunodeficiency | Meningoencephalitis | None | Not described | Dead | [41] |
| Mamastrovirus 6 | |||||||||
| MLB1 | 2015 | Japan | 4 years | CB HSCT for congenital aplastic anemia GvH disease | Encephalitis | Aciclovir IVIG Edaravone | Ciclosporin MMF | Alive | [53] |
| MLB2 | 2014 | Switzerland | 21 years | Healthy | Acute meningitis | Ceftriaxone Aciclovir | None | Alive | [52] |
| MLB2 | 2014 | Switzerland | 37 years | HSCT for acute myeloid leukemia, relapse | Meningitis | None | IT chemotherapy 5-AZC Cranial irradiation | Dead | [52] |
| Mamastrovirus 9 | |||||||||
| VA1 (HAstV-PS) | 2007 | US | 15 years | X-linked agammaglobulinemia | Headache, suicidal and homicidal ideation, memory loss, ataxia, progressive cognitive decline | None | Related to underlying disease | Dead | [47] |
| VA1 | 2013 | UK | 42 years | HSCT for chronic lymphocytic leukemia | Progressive sensorineural deafness Encephalitis | Valaciclovir BS antibiotics Steroids IVIG Ribavirin | Not described | Dead | [49] |
| VA1 | 2014 | France | 14 years | X-linked agammaglobulinemia | Four-year history of progressive cognitive impairment, ataxia and seizure. | IVIG Steroid Ribavirin PEG IFN alpha-2b | Related to underlying disease | Alive | [50] |
| VA1 | 2015 | UK | 18 months | HSCT for cartilage hair hypoplasia GvH disease | Encephalitis | Cidofovir * Adenovirus-specific DLI * | Ciclosporin MMF Steroids | Dead | [48] |
| VA1 | 2015 | UK | 8 months | HSCT for acute myeloid leukemia GvH grade 1 | Encephalitis | DLI | Ciclosporin ** | Dead | [51] |
| Geographical Area (Time of Study) | Type of Individuals | Method | Positivity Rate (%) | % of Positive Samples Containing Other Pathogens (Type) | Serotype Prevalences | Reference |
|---|---|---|---|---|---|---|
| Children with symptoms of AGE | ||||||
| Asia | ||||||
| China (2007–2008) | Outpatients < 15 | RT-PCR | 13.6 | N/A | HAstV-1 (100%) | [76] |
| China (2008–2009) | Hospitalized < 5 | RT-PCR | 4.6 | 26 (other enteric viruses) | HAstV-1 (100%) | [77] |
| China (2010–2011) | Outpatients < 5 | RT-PCR | 1.8 | 50 (other enteric viruses) | HAstV-1 (100%) | [78] |
| China (2010–2011) | Outpatients < 5 | RT-PCR | 2.9 | 64 (rotavirus) | HAstV-1 (100%) | [68] |
| China (2005–2006) | <5 | RTqPCR | 9.1 | N/A | HAstV-1 (96%); HAstv-3 (4%) | [79] |
| India (2004–2008) | Hospitalized < 5 | RT-PCR | 3.1 | 8.8 (rotavirus) | HAstV-1 (68%); HAstv-2 (10%); HAstV-8 (16%); HAstV-5 (6%) | [80] |
| Japan (2009–2013) | Outpatients < 5 | RT-PCR | 2.4 | 0 | N/A | [81] |
| Japan (2009/10, 2014/15) | <15 | RT-PCR | 4.2 | N/A | HAstV-1 (54%); HAstv-4 (23%); HAstV-8 (16%); HAstV-6 (7%) | [82] |
| Japan (2008/09, 2013/14) | Hospitalized < 15 with suspected viral gastroenteritis | RT-PCR | 1.6 | N/A | HAstV-1 (81%); HAstV-8 (16%); HAstV-3 (3%) | [83] |
| Japan (2012–2013) | Outpatients | RT-PCR | 5.2 | 29 (other enteric viruses) | HAstV-1 (76%); HAstv-4 (24%) | [84] |
| Taiwan (2009–2011) | Hospitalized < 5 | RT-PCR | 2.6 | 20 (other enteric viruses) | N/A | [85] |
| Thailand (2000–2003, 2005, 2007–2008, 2010–2011) | Hospitalized < 5 | RT-PCR | 1.4 | 14 (rotavirus) | HAstV-1 (58%); HAstv-3 (21%); HAstV-5 (14%); HAstV-3 (7%) | [86] |
| Vietnam (2002–2003) | Hospitalized < 9 | Multiplex RT-PCR | 0.6 | 33 (other enteric viruses) | HAstV-1 (100%) | [72] |
| Vietnam (2005–2006) | Hospitalized and outpatients < 15 | RT-PCR | 13.9 | 28 (other enteric viruses) | HAstV-1 (100%) | [87] |
| Africa | ||||||
| Burkina Faso (November 2011–September 2012) | Outpatients < 5 | RTqPCR | 4.9 | 7.7 (other enteric viruses) | HAstV-1 (42%); HAstv-2 (25%); HAstV-8 (25%); HAstV-5 (8%) | [88] * |
| Gabon (2010–2011) | Outpatients < 5 | RT-PCR | 6.3 | 55 (other enteric viruses) | HAstV-1 (89%); HAstv-4 (11%) | [89] |
| Ghana (November 2005–January 2006) | Outpatients < 5 | RT-PCR | 4.8 | N/A | N/A | [90] * |
| Kenya and Gambia (2008–2009) | < 5 | RT-PCR | 2.7 | N/A | N/A | [20] * |
| Europe and Middle East | ||||||
| Bulgaria (summer 2009) | Hospitalized < 3, summer months | RT-PCR | 6.9 | 50 (other enteric viruses, bacteria and parasites) | HAstV-1 (86%); HAstv-3 (14%) | [91] |
| Finland (2009–2010) | Children < 2 enrolled in prospective cohort INDIS Study | RTqPCR | 1.9 | 33 (other enteric viruses) | N/A | [92] |
| Italy (2008–2009) | Hospitalized < 13 | RT-PCR | 2.1 | 0 (other enteric viruses) | HAstV-1 (73%); HAstv-2 (20%); HAstV-4 (7%) | [93] |
| Italy (2008–2009) | Hospitalized < 18 | Multiplex RT-PCR | 0 | 0 | N/A | [73] |
| Moldova and Ukraine (2009) | Hospitalized < 5, negative for rotavirus | RTqPCR | 1.4 | 14.3 (other enteric viruses) | HAstV-1 (80%); HAstv-8 (20%) | [94] |
| Qatar (June-November 2009) | Outpatients < 20 | Multiplex RTqPCR | 0.7 | N/A | N/A | [74] |
| United Kingdom (2006–2007) | Hospitalized < 16, health-care associated AGE | RT-PCR | 5 | 57 (other enteric viruses) | N/A | [71] |
| Central and South America | ||||||
| Brazil (1994–1996; 1995–1999) | Outpatients < 6 | RT-PCR | 7.6; 29.7 | 22; 50 children with AGE and controls (other enteric viruses) | HAstV-1 (58%); HAstV-2 (24%); HAstV-8 (12%); HAstV-3 (6%) | [38] * |
| Brazil (1997–1999) | Outpatients < 2 | RT-PCR | 11 | 55 (other enteric viruses) | HAstV-1 (92%); HAstV-2 (2%); HAstV-3 (2%); HAstV-4 (2%); HAstV-5 (2%) | [95] * |
| Brazil (1994–1996; 1998–2002) | Hospitalized < 5 | RT-PCR | 4.3 | 30.4 children with AGE and controls (other enteric viruses) | N/A | [96] * |
| Brazil (2005–2011) | Children < 5, negative for rotavirus and norovirus | RT-PCR | 7.1 | N/A | HAstV-1 (70%); HAstV-2 (12%); HAstV-3 (10%); HAstV-8 (4%); HAstV-4 (2%); HAstV-6 (2%) | [69] |
| Venezuela (2003) | Outpatients < 5 | Multiplex RT-PCR | 1.5 | 29 (other enteric viruses) | HAstV-1 (67%); HAstV-3 (33%) | [75] |
| North America | ||||||
| US (2006–2009) | Hospitalized and outpatients | RT-PCR | 3.1 | N/A | N/A | [8] |
| US (2008–2009) | Hospitalized and outpatients < 5 | RTqPCR | 4.9 | 25 children with AGE and controls (other enteric viruses) | HAstV-1 (52%); HAstV-2 (19%); HAstV-4 (8%); HAstV-8 (3%) | [39] * |
| Children without diarrhea disorders | ||||||
| Burkina Faso (November 2011–September 2012) | Matched controls < 5 | RTqPCR | 2 | N/A | HAstV-1 (42%); HAstv-2 (25%); HAstV-8 (25%); HAstV-5 (8%) | [88] * |
| Brazil (1997–1999) | < 2 | RT-PCR | 3 | 20 (other enteric viruses) | HAstV-1 (92%); HAstV-2 (2%); HAstV-3 (2%); HAstV-4 (2%); HAstV-5 (2%) | [95] * |
| Brazil (1994–1996; 1995–1999) | < 6 | RT-PCR | 20.7; 16.3 | 22; 50 children with AGE and controls (other enteric viruses) | HAstV-1 (58%); HAstV-2 (24%); HAstV-8 (12%); HAstV-3 (6%) | [38] * |
| Brazil (1994–1996; 1998–2002) | < 5 | RT-PCR | 0.5 | 30.4 children with AGE and controls (other enteric viruses) | N/A | [96] * |
| Ghana (November 2005–January 2006) | Matched controls < 5 | RT-PCR | 1.6 | N/A | N/A | [90] * |
| Kenya and Gambia (2008–2009) | < 5 | RT-PCR | 2.4 | N/A | N/A | [20] * |
| US (2008–2009) | Matched controls < 5 | RTqPCR | 3.0 | 25 children with AGE and controls (other enteric viruses) | HAstV-3 (57%) | [39] * |
| Adults with AGE | ||||||
| China (2005–2006) | Collected from CDC’s surveillance | RTqPCR | 5.4 | N/A | HAstV-1 (96%); HAstv-3 (4%) | [79] |
| China (2007–2008) | Visiting an outpatient clinic and/or emergency room | RT-PCR | 1.8 | 30 (other enteric viruses) | N/A | [97] |
| France (2010–2011) | Consulting a general practitioner | RT-PCR | 6.9 | 50 (other enteric viruses) | N/A | [98] |
| Russia (2005–2007) | Hospitalized | RT-PCR | 2.2 | N/A | N/A | [99] |
| Singapore (October 2013–January 2014) | Hospitalized | RT-PCR | 2 | N/A | N/A | [100] |
| US (2006–2009) | Hospitalized and outpatients | RT-PCR | 1.2 | N/A | N/A | [8] |
| Immunocompromised | ||||||
| Brazil (2003–2004) | HIV-seropositive children with and without diarrhea | RT-PCR | 0; 11 | 0 | N/A | [7] |
| US (2006–2009) | Hospitalized | RT-PCR | 7.4 | N/A | N/A | [8] |
| Geographical Area (Time of Study) | Type of Individuals | Method | MLB Positivity Rate (%) | VA Positivity Rate (%) | Reference |
|---|---|---|---|---|---|
| Children and adults with AGE | |||||
| India (2005–2006) | Community-based samples from a birth cohort | RT-PCR | 2.1 | 0.7 | [12] |
| China (2004–2005) | Hospitalized patients | RT-PCR | 0.2 | 0 | [116] |
| China (2010–2011) | Outpatients < 5 | RT-PCR | 1.2 | 0.3 | [68] |
| Japan (2012–2013) | Outpatient children | RT-PCR | 10.6 | 0.6 | [84] |
| Nepal (2006–2008) | Adults, negative for bacteria, rotavirus, adenovirus, HAstV, Giardia, Cryptosporidium or norovirus. | RT-PCR | 0 | 2.1 | [14] * |
| Kenya and Gambia (2008–2009) | Children < 5 from rural areas | RT-PCR | 4.3 | 1.3 | [20] * |
| UK (2013–2014) | Immunosuppressed and immunocompetent children and adults | RTqPCR (VA1) | N/A | 0.3 | [48] |
| Egypt (2006–2007) | Outpatients < 5 | RT-PCR | 1.4 | 0.5 | [117] |
| Turkey (2004–2005) | Children < 5, negative for rotavirus | RT-PCR | 0.7 | 0 | [118] |
| Brazil (2005–2011) | Children < 2, negative for rotavirus and norovirus | RT-PCR | 1 | 0 | [69] |
| US (2008) | < 5 | RT-PCR | 0.6 | 0 | [13] |
| Children and adults without diarrhea disorders | |||||
| Nepal (2006–2008) | Adults, negative for bacteria, rotavirus, adenovirus, HAstV, Giardia, Cryptosporidium or norovirus | RT-PCR | 0 | 1 | [14] * |
| Kenya and Gambia (2008–2009) | Children < 5 from rural areas | RT-PCR | 6.4 | 1.8 | [20] * |
| Children with non-polio acute flaccid paralysis (AFP) | |||||
| Nigeria (2006–2008) | Children < 15 | RT-PCR | 4.2 | 3.2 | [14] |
| Pakistan (2006–2008) | Children < 15 | RT-PCR | 0 | 4.6 | [14] |
| Children and adults (undefined clinical presentation) | |||||
| Switzerland (2014–2015) | Children and adults stool specimens stored at a Laboratory of Virology of a University hospital | RTqPCR (MLB2) | 0.9 | N/A | [52] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vu, D.-L.; Bosch, A.; Pintó, R.M.; Guix, S. Epidemiology of Classic and Novel Human Astrovirus: Gastroenteritis and Beyond. Viruses 2017, 9, 33. https://doi.org/10.3390/v9020033
Vu D-L, Bosch A, Pintó RM, Guix S. Epidemiology of Classic and Novel Human Astrovirus: Gastroenteritis and Beyond. Viruses. 2017; 9(2):33. https://doi.org/10.3390/v9020033
Chicago/Turabian StyleVu, Diem-Lan, Albert Bosch, Rosa M. Pintó, and Susana Guix. 2017. "Epidemiology of Classic and Novel Human Astrovirus: Gastroenteritis and Beyond" Viruses 9, no. 2: 33. https://doi.org/10.3390/v9020033
APA StyleVu, D. -L., Bosch, A., Pintó, R. M., & Guix, S. (2017). Epidemiology of Classic and Novel Human Astrovirus: Gastroenteritis and Beyond. Viruses, 9(2), 33. https://doi.org/10.3390/v9020033








