Advanced Microfluidic Technologies for Lipid Nano-Microsystems from Synthesis to Biological Application
Abstract
:1. Introduction
1.1. Overview of Microfluidic Techniques for Lipid Nano-Micro Sized Structures
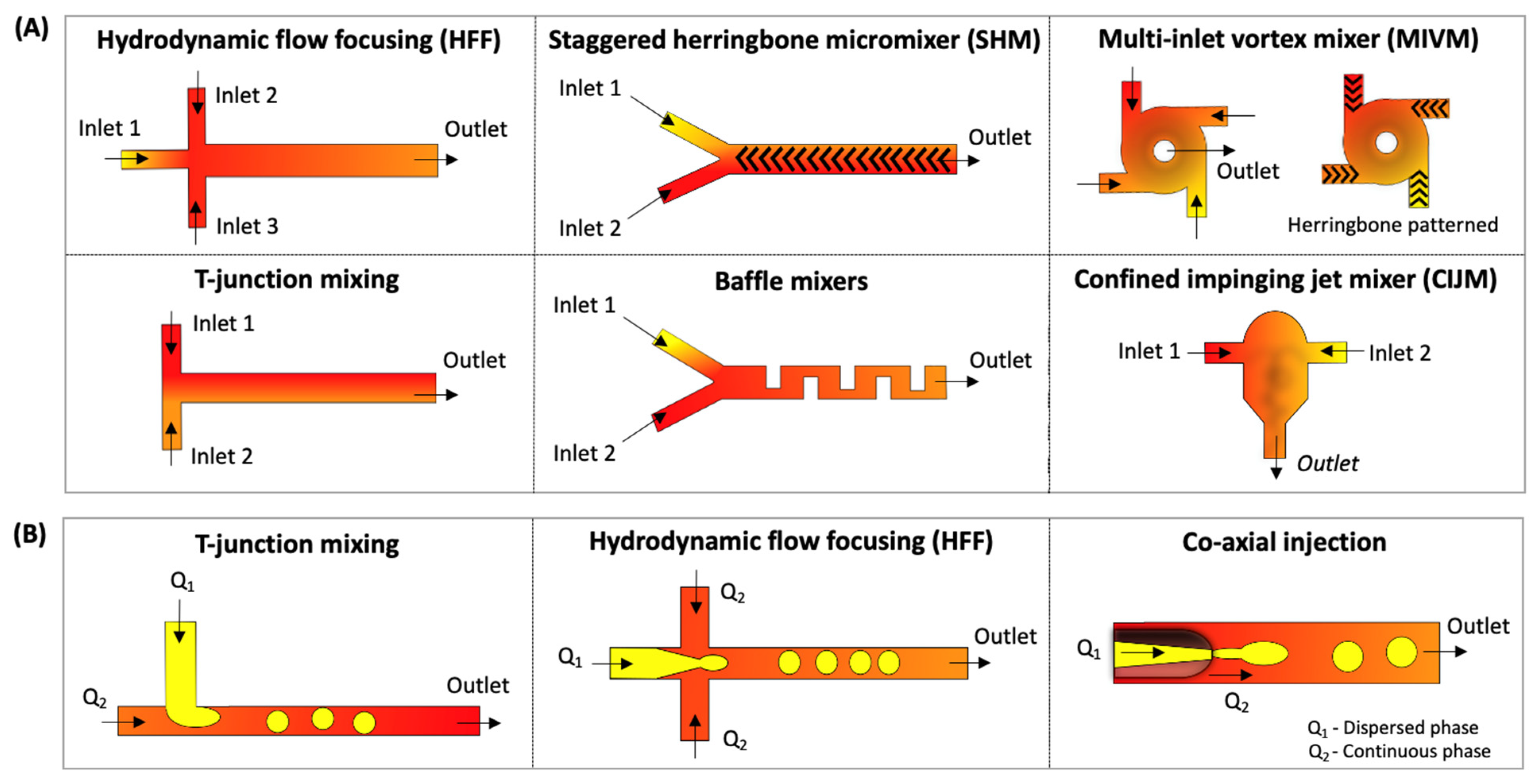
1.1.1. Microfluidic Hydrodynamic Flow-Focusing (HFF)
1.1.2. Microfluidic Chaotic Advection Micromixers (CA-M)
1.1.3. Droplet-Based Microfluidics
2. Microfluidics for Lipid Nano-Sized Structures Synthesis: Drug and Gene Delivery
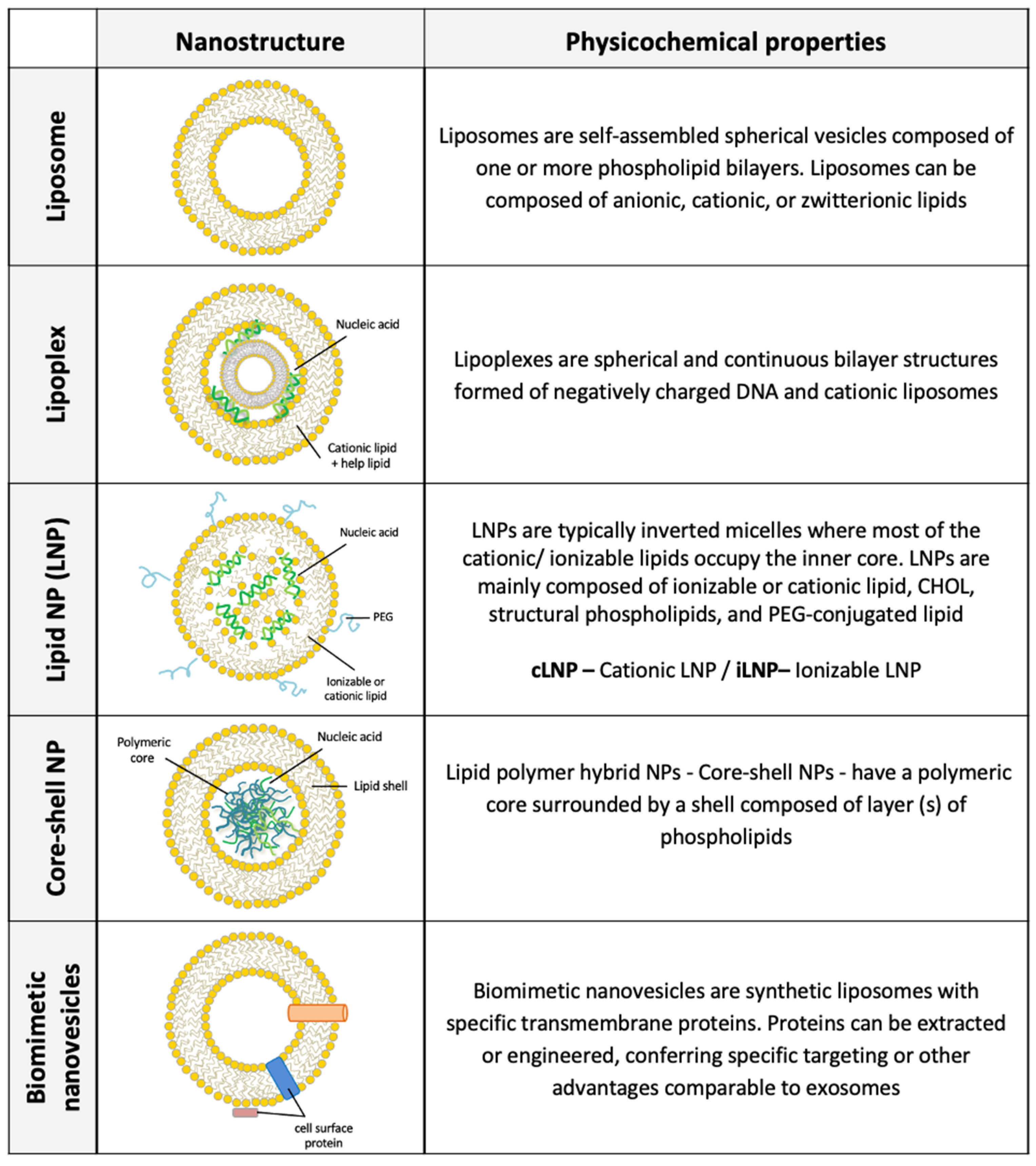
2.1. Liposomes for Drug Delivery
2.2. Cationic Liposomes (CLs) and Cationic LNPs for Gene Delivery
2.3. pDNA-Based Non-Viral Lipid Vectors
2.4. RNA-Based Non-Viral Lipid Vectors
2.5. Lipid-Polymer Hybrid (Core/Shell) Nanoparticles Synthesis for Drug/Gene Delivery
2.6. Biomimetic Nanovesicles
3. Microfluidics for Lipid Micro-Sized Structures Synthesis
4. Additional Approaches for Sustained Release of Liposomes and Screening of Lipid Nanostructure Using Microfluidics
4.1. Microencapsulation of Liposomes for Drug Delivery Using Droplet-Based Microfluidics
4.2. Microfluidic Platforms for Lipid-Based Nanocarriers Assays (Drug/Gene Delivery)
5. Future Perspectives and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DOPE | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine |
| PCL-PEI | polyethylenimine-graft-polycaprolactone |
| CHOL | cholesterol |
| DSPE-PEG2000 | 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy (polyethylene glycol)]-2000 |
| PEI | polyethylenimine |
| DPPC | 1,2-dipalmitoyl-sn-glycero-3- phosphocholine |
| DMG-PEG | 1,2-dimyristoyl-rac-glycero-3-methoxy poly (ethylene glycol)-2000 |
| PLGA | poly (lactic-co- glycolic acid) |
| DOTAP | 1,2-dioleoyl-3-trimethylammonium-propane |
| CPP-SA | poly-carboxyphenoxy propane co-sebacic acid |
| PEG | polyethylene glycol |
| DC-Chol | 3-β-[N-(N0, N0-dimethylaminoethane)-carbamoyl])-cholesterol |
| DOPC | 1,2-dioleoyl-sn-glycero-3-phosphocholine |
| EPC | egg phosphatidylcholine |
| DSPC | 1,2-distearoyl-sn-glycero-3-phosphocholine |
| PC | phosphatidylcholine |
| DMPC | 1,2-dimyristoyl-sn-gly- cero-3-phosphocholine |
| DPPC | 1,2-dipalmitoyl-sn-glycero-3-phos-phocholine |
| PS | l-α-phosphatidylserine |
| HSPC | hydrogenated soy l- α-phosphatidylcholine (HSPC) |
| KC2 | 2,2-dilinoleyl-4-(2-dimethylaminoethyl)-[1,3]-dioxolane |
| PEG-DMPE | 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] |
| DOPE-PEG | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)]-2000 |
| DLinDAP | 1,2-dilinoleoyl-3-dimethylaminopropane |
| DLinDMA | 1,2-dilinoleyloxy-3-dimethylaminopropane |
| MC3 | heptatriaconta-6,9,28,31-tetraen-19-yl 4-(dimethylamino)butanoate |
| SOPC | 1-stearoyl-2-oleoyl-sn-glycero-3-phosphocholine |
| DDAB | dimethyldioctadecylammonium |
| DODMA | 1,2-dioleyloxy-3-dimethylaminopropane |
| DOBAQ | N-(4-carboxybenzyl)-N,N-dimethyl-2,3-bis(oleoyloxy)propan-1-aminium |
| DOTMA | 1,2-di-O-octadecenyl-3-trimethylammonium propane |
| DDA | dimethyldioctadecylammonium |
| DMTAP | 1,2-dimyristoyl-3-trimethylammonium-propane |
| DSTAP | 1,2-stearoyl-3-trimethylammonium-propane |
| DLinPC | 1,2-dilinoleoyl-sn-glycero-3-phosphorylcholine |
| DPoPC | 1-palmitoyl,2-oleoyl-sn-glycero-3-phosphorylcholine |
| TPGS | d-α-tocopherol polyethylene glycol 1000 succinate |
| DAP | 1,2-dipalmitoyl-3-dimethylammonium-propane |
| HFF | hydrodynamic flow-focusing |
| CA-M | chaotic advection-based micromixer |
| SHM | staggered herringbone micromixer |
| MIVM | multi-inlet vortex mixer |
| CIJM | confined impingement jet mixer |
References
- Bangham, A.D.; Standish, M.M.; Watkins, J.C. Diffusion of univalent ions across the lamellae of swollen phospholipids. J. Mol. Biol. 1965, 13, 238–252. [Google Scholar] [CrossRef]
- Fan, Y.; Zhang, Q. Development of liposomal formulations: From concept to clinical investigations. Asian J. Pharm. Sci. 2013, 8, 81–87. [Google Scholar] [CrossRef] [Green Version]
- Pattni, B.S.; Chupin, V.V.; Torchilin, V.P. New Developments in Liposomal Drug Delivery. Chem. Rev. 2015, 115, 10938–10966. [Google Scholar] [CrossRef]
- Lasic, D.D. The mechanism of vesicle formation. Biochem. J. 1988, 256, 1–11. [Google Scholar] [CrossRef]
- Bozzuto, G.; Molinari, A. Liposomes as nanomedical devices. Int. J. Nanomed. 2015, 975–999. [Google Scholar] [CrossRef] [Green Version]
- Phapal, S.M.; Sunthar, P. Influence of micro-mixing on the size of liposomes self-assembled from miscible liquid phases. Chem. Phys. Lipids 2013, 172–173, 20–30. [Google Scholar] [CrossRef] [Green Version]
- Filipczak, N.; Pan, J.; Yalamarty, S.S.K.; Torchilin, V.P. Recent advancements in liposome technology. Adv. Drug Deliv. Rev. 2020, 156, 4–22. [Google Scholar] [CrossRef]
- Carugo, D.; Bottaro, E.; Owen, J.; Stride, E.; Nastruzzi, C. Liposome production by microfluidics: Potential and limiting factors. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Barenholz, Y. Doxil®—The first FDA-approved nano-drug: Lessons learned. J. Control. Release 2012, 160, 117–134. [Google Scholar] [CrossRef]
- Allen, T.M.; Martin, F.J. Advantages of liposomal delivery systems for anthracyclines. Semin. Oncol. 2004, 31, 5–15. [Google Scholar] [CrossRef]
- Maeki, M.; Kimura, N.; Sato, Y.; Harashima, H.; Tokeshi, M. Advances in microfluidics for lipid nanoparticles and extracellular vesicles and applications in drug delivery systems. Adv. Drug Deliv. Rev. 2018, 128, 84–100. [Google Scholar] [CrossRef]
- Ickenstein, L.M.; Garidel, P. Lipid-based nanoparticle formulations for small molecules and RNA drugs. Expert Opin. Drug Deliv. 2019, 16, 1205–1226. [Google Scholar] [CrossRef] [PubMed]
- Dunbar, C.E.; High, K.A.; Joung, J.K.; Kohn, D.B.; Ozawa, K.; Sadelain, M. Gene therapy comes of age. Science 2018, 359, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Verma, I.M.; Naldini, L.; Kafri, T.; Miyoshi, H.; Takahashi, M.; Blomer, U.; Somia, N. Gene therapy—Promises, problems and prospects. Genes Resist. Dis. 2000, 389, 147–157. [Google Scholar]
- Jeong, J.H.; Kim, S.W.; Park, T.G. Molecular design of functional polymers for gene therapy. Prog. Polym. Sci. 2007, 32, 1239–1274. [Google Scholar] [CrossRef]
- Edelstein, M.L.; Abedi, M.R.; Wixon, J. Gene therapy clinical trials worldwide to 2007—An update. J. Gene Med. 2007, 9, 833–842. [Google Scholar] [CrossRef]
- Del Pozo-Rodríguez, A.; Solinís, M.Á.; Rodríguez-Gascón, A. Applications of lipid nanoparticles in gene therapy. Eur. J. Pharm. Biopharm. 2016. [Google Scholar] [CrossRef]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Glover, D.J.; Lipps, H.J.; Jans, D.A. Towards safe, non-viral therapeutic gene expression in humans. Nat. Rev. Genet. 2005, 6, 299–310. [Google Scholar] [CrossRef]
- Neergaard, L.; Perrone, M. US Regulators Give Full Approval to Pfizer COVID-19 Vaccine. Available online: https://apnews.com/article/coronavirus-vaccine-pfizer-approval-1361ff61d06b815652a08a7cc0683a72 (accessed on 25 August 2021).
- Shepherd, S.J.; Warzecha, C.C.; Yadavali, S.; El-Mayta, R.; Alameh, M.G.; Wang, L.; Weissman, D.; Wilson, J.M.; Issadore, D.; Mitchell, M.J. Scalable mRNA and siRNA Lipid Nanoparticle Production Using a Parallelized Microfluidic Device. Nano Lett. 2021, 21, 5671–5680. [Google Scholar] [CrossRef] [PubMed]
- Aldosari, B.N.; Alfagih, I.M.; Almurshedi, A.S. Lipid nanoparticles as delivery systems for RNA-based vaccines. Pharmaceutics 2021, 13, 206. [Google Scholar] [CrossRef]
- Justo, O.R.; Moraes, Â.M. Analysis of process parameters on the characteristics of liposomes prepared by ethanol injection with a view to process scale-up: Effect of temperature and batch volume. Chem. Eng. Res. Des. 2011, 89, 785–792. [Google Scholar] [CrossRef]
- Lo, R. Application of Microfluidics in Chemical Engineering. Chem. Eng. Process Tech. 2013, 1002. [Google Scholar]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef]
- Fang, A.; Cathala, B. Smart swelling biopolymer microparticles by a microfluidic approach: Synthesis, in situ encapsulation and controlled release. Colloids Surf. B Biointerfaces 2011, 82, 81–86. [Google Scholar] [CrossRef]
- Ushikubo, F.Y.; Oliveira, D.R.B.; Michelon, M.; Cunha, R.L. Designing Food Structure Using Microfluidics. Food Eng. Rev. 2015, 7, 393–416. [Google Scholar] [CrossRef]
- BCC Publishing. Microfluidics: Technologies and Global Markets. Available online: https://www.bccresearch.com/market-research/semiconductor-manufacturing/microfluidics-technologies-and-global-markets.html (accessed on 17 November 2021).
- Utada, A.S.; Lorenceau, E.; Link, D.R.; Kaplan, P.D.; Stone, H.A.; Weitz, D.A. Monodisperse double emulsions generated from a microcapillary device. Science 2005, 308, 537–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atencia, J.; Beebe, D.J. Controlled microfluidic interfaces. Nature 2005, 437, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Zhigaltsev, I.V.; Belliveau, N.; Hafez, I.; Leung, A.K.K.; Huft, J.; Hansen, C.; Cullis, P.R. Bottom-up design and synthesis of limit size lipid nanoparticle systems with aqueous and triglyceride cores using millisecond microfluidic mixing. Langmuir 2012, 28, 3633–3640. [Google Scholar] [CrossRef] [PubMed]
- Webb, C.; Forbes, N.; Roces, C.B.; Anderluzzi, G.; Lou, G.; Abraham, S.; Ingalls, L.; Marshall, K.; Leaver, T.J.; Watts, J.A.; et al. Using microfluidics for scalable manufacturing of nanomedicines from bench to GMP: A case study using protein-loaded liposomes. Int. J. Pharm. 2020, 582, 119266. [Google Scholar] [CrossRef] [PubMed]
- Batzri, S.; Korn, E.D. Single bilayer liposomes prepared without sonication. BBA Biomembr. 1973, 298, 1015–1019. [Google Scholar] [CrossRef]
- Convery, N.; Gadegaard, N. 30 Years of Microfluidics. Micro Nano Eng. 2019, 2, 76–91. [Google Scholar] [CrossRef]
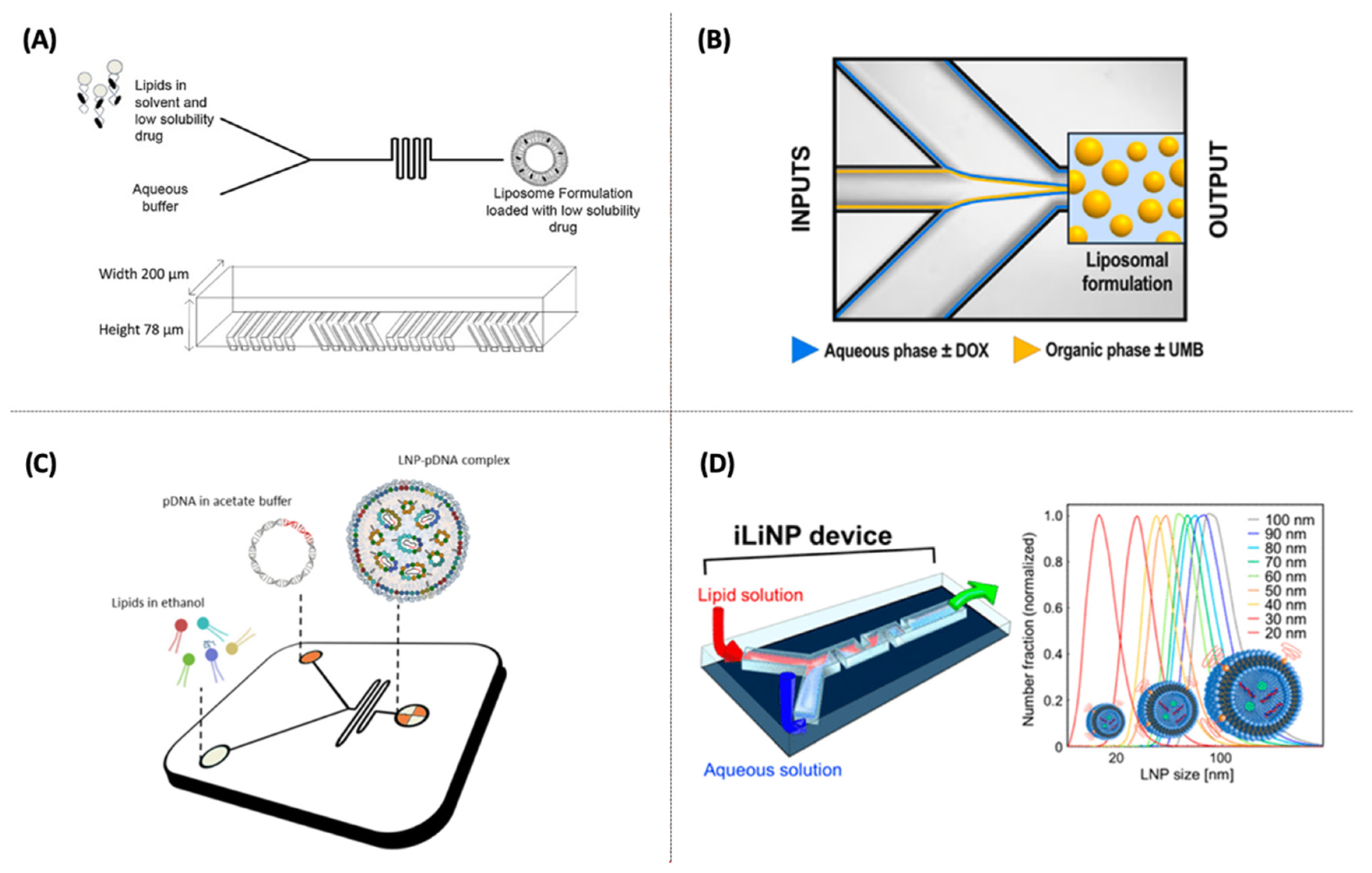
- Michelon, M.; Oliveira, D.R.B.; de Figueiredo Furtado, G.; de la Torre, L.G.; Cunha, R.L. High-throughput continuous production of liposomes using hydrodynamic flow-focusing microfluidic devices. Colloids Surf. B Biointerfaces 2017, 156, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Jahn, A.; Reiner, J.E.; Vreeland, W.N.; DeVoe, D.L.; Locascio, L.E.; Gaitan, M. Preparation of nanoparticles by continuous-flow microfluidics. J. Nanoparticle Res. 2008, 10, 925–934. [Google Scholar] [CrossRef]
- Amrani, S.; Tabrizian, M. Characterization of Nanoscale Loaded Liposomes Produced by 2D Hydrodynamic Flow Focusing. ACS Biomater. Sci. Eng. 2018, 4, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Aghaei, H.; Solaimany Nazar, A.R. Continuous Production of the Nanoscale Liposome in a Double Flow-Focusing Microfluidic Device. Ind. Eng. Chem. Res. 2019, 58, 23032–23045. [Google Scholar] [CrossRef]
- Kumar, S.L. Microfluidics technology for nanoparticles and equipment. In Emerging Technologies for Nanoparticle Manufacturing; Patel, J.K., Pathak, Y.V., Eds.; Springer Nature: Cham, Switzerland, 2021; pp. 67–98. ISBN 9783030507022. [Google Scholar]
- Lee, J.N.; Park, C.; Whitesides, G.M. Solvent Compatibility of Poly(dimethylsiloxane)-Based Microfluidic Devices. Anal. Chem. 2003, 75, 6544–6554. [Google Scholar] [CrossRef]
- Sollier, E.; Murray, C.; Maoddi, P.; Di Carlo, D. Rapid prototyping polymers for microfluidic devices and high pressure injections. Lab Chip 2011, 11, 3752–3765. [Google Scholar] [CrossRef]
- Han, T.; Zhang, L.; Xu, H.; Xuan, J. Factory-on-chip: Modularised microfluidic reactors for continuous mass production of functional materials. Chem. Eng. J. 2017, 326, 765–773. [Google Scholar] [CrossRef]
- Jeong, H.H.; Issadore, D.; Lee, D. Recent developments in scale-up of microfluidic emulsion generation via parallelization. Korean J. Chem. Eng. 2016, 33, 1757–1766. [Google Scholar] [CrossRef]
- You, J.B.; Kang, K.; Tran, T.T.; Park, H.; Hwang, W.R.; Kim, J.M.; Im, S.G. PDMS-based turbulent microfluidic mixer. Lab Chip 2015, 15, 1727–1735. [Google Scholar] [CrossRef]
- Halldorsson, S.; Lucumi, E.; Gómez-Sjöberg, R.; Fleming, R.M.T. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens. Bioelectron. 2015, 63, 218–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, K.I.N.F. Materials and Fabrication Techniques for Nano- and Microfluidic Devices. In Microfluidics in Detection Science: Lab-on-a-Chip Technologies; Labeed, F.H., Fatoyinbo, H.O., Eds.; Royal Society of Chemistry: Washington, DC, USA, 2015; pp. 1–28. ISBN 9781849737609. [Google Scholar]
- Michelon, M.; Huang, Y.; de la Torre, L.G.; Weitz, D.A.; Cunha, R.L. Single-step microfluidic production of W/O/W double emulsions as templates for Β-carotene-loaded giant liposomes formation. Chem. Eng. J. 2019, 366, 27–32. [Google Scholar] [CrossRef]
- Schmitt, P.; Wedrich, K.; Müller, L.; Mehner, H.; Hoffmann, M. Design, fabrication and characterisation of a microfluidic time-temperature indicator. J. Phys. Conf. Ser. 2017, 922, 012004. [Google Scholar] [CrossRef] [Green Version]
- Martinez, A.W.; Phillips, S.T.; Whitesides, G.M. Diagnostics for the Developing World: Microfluidic Paper-Based Analytical Devices. Anal. Chem. 2010, 82, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Jafry, A.T.; Lim, H.; Sung, W.K.; Lee, J. Flexible time–temperature indicator: A versatile platform for laminated paper-based analytical devices. Microfluid. Nanofluid. 2017, 21, 57. [Google Scholar] [CrossRef]
- Eş, I.; Montebugnoli, L.J.; Filippi, M.F.P.; Malfatti-Gasperini, A.A.; Radaic, A.; Bispo, M.; de Jesus, M.B.; De La Torre, L.G. High-throughput conventional and stealth cationic liposome synthesis using a chaotic advection-based microfluidic device combined with a centrifugal vacuum concentrator. Chem. Eng. J. 2020, 382, 122821. [Google Scholar] [CrossRef]
- Beebe, D.J.; Mensing, G.A.; Walker, G.M. Physics and applications of microfluidics in biology. Annu. Rev. Biomed. Eng. 2002, 4, 261–286. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Whitesides, G.M. Soft lithography. Annu. Rev. Mater. Sci. 1998. [Google Scholar] [CrossRef]
- Dungchai, W.; Chailapakul, O.; Henry, C.S. A low-cost, simple, and rapid fabrication method for paper-based microfluidics using wax screen-printing. Analyst 2011, 136, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Sameenoi, Y.; Na Nongkai, P.; Nouanthavong, S.; Henry, C.S.; Nacaprich, D. One-step polymer screen-printing for microfluidic paper-based analytical device (uPAD) fabrication. Analyst 2014, 139, 6580–6588. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wu, C.; Chu, P.K.; Gelinsky, M. 3D printing of hydrogels: Rational design strategies and emerging biomedical applications. Mater. Sci. Eng. R 2020, 140, 100543. [Google Scholar] [CrossRef]
- Highley, C.B.; Rodell, C.B.; Burdick, J.A. Direct 3D Printing of Shear-Thinning Hydrogels into Self- Healing Hydrogels. Adv. Mater. 2015, 27, 5075–5079. [Google Scholar] [CrossRef] [PubMed]
- Dimov, N.; Kastner, E.; Hussain, M.; Perrie, Y.; Szita, N. Formation and purification of tailored liposomes for drug delivery using a module-based micro continuous-flow system. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Charmet, J.; Rodrigues, R.; Yildirim, E.; Challa, P.K.; Roberts, B.; Dallmann, R.; Whulanza, Y. Low-Cost microfabrication tool box. Micromachines 2020, 11, 135. [Google Scholar] [CrossRef] [Green Version]
- Ling, F.W.M.; Mahmood, W.K.; Abdulbari, H.A. Rapid Prototyping of Microfluidics Devices using Xurograhy Method. MATEC Web Conf. 2017, 111, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Abe, K.; Suzuki, K.; Citterio, D. Inkjet-Printed Microfluidic Multianalyte Chemical Sensing Paper. Anal. Chem. 2008, 80, 6928–6934. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.F.; Pessoa, A.C.S.N.; Bastos, R.G.; de la Torre, L.G. Microfluidic tools toward industrial biotechnology. Biotechnol. Prog. 2016, 32, 1372–1389. [Google Scholar] [CrossRef]
- Ren, K.; Zhou, J.; Wu, H. Materials for microfluidic chip fabrication. Account. Chem. Res. 2013, 46, 2396–2406. [Google Scholar] [CrossRef] [PubMed]
- Jahn, A.; Vreeland, W.N.; Gaitan, M.; Locascio, L.E. Controlled Vesicle Self-Assembly in Microfluidic Channels with Hydrodynamic Focusing. J. Am. Chem. Soc. 2004, 126, 2674–2675. [Google Scholar] [CrossRef]
- Shepherd, S.J.; Issadore, D.; Mitchell, M.J. Microfluidic formulation of nanoparticles for biomedical applications. Biomaterials 2021, 274, 120826. [Google Scholar] [CrossRef]
- Evers, M.J.W.; Kulkarni, J.A.; van der Meel, R.; Cullis, P.R.; Vader, P.; Schiffelers, R.M. State-of-the-Art Design and Rapid-Mixing Production Techniques of Lipid Nanoparticles for Nucleic Acid Delivery. Small Methods 2018, 2, 1700375. [Google Scholar] [CrossRef]
- Ahn, J.; Ko, J.; Lee, S.; Yu, J.; Kim, Y.T.; Jeon, N.L. Microfluidics in nanoparticle drug delivery; From synthesis to pre-clinical screening. Adv. Drug Deliv. Rev. 2018, 128, 29–53. [Google Scholar] [CrossRef] [PubMed]
- Kastner, E.; Verma, V.; Lowry, D.; Perrie, Y. Microfluidic-controlled manufacture of liposomes for the solubilisation of a poorly water soluble drug. Int. J. Pharm. 2015, 485, 122–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forbes, N.; Hussain, M.T.; Briuglia, M.L.; Edwards, D.P.; Ter Horst, J.H.; Szita, N.; Perrie, Y. Rapid and scale-independent microfluidic manufacture of liposomes entrapping protein incorporating in-line purification and at-line size monitoring. Int. J. Pharm. 2019, 556, 68–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roces, C.B.; Lou, G.; Jain, N.; Abraham, S.; Thomas, A.; Halbert, G.W.; Perrie, Y. Manufacturing considerations for the development of lipid nanoparticles using microfluidics. Pharmaceutics 2020, 12, 1095. [Google Scholar] [CrossRef] [PubMed]
- Sago, C.D.; Lokugamage, M.P.; Paunovska, K.; Vanover, D.A.; Monaco, C.M.; Shah, N.N.; Castro, M.G.; Anderson, S.E.; Rudoltz, T.G.; Lando, G.N.; et al. High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc. Natl. Acad. Sci. USA 2018, 115. [Google Scholar] [CrossRef] [Green Version]
- Lou, G.; Anderluzzi, G.; Schmidt, S.T.; Woods, S.; Gallorini, S.; Brazzoli, M.; Giusti, F.; Ferlenghi, I.; Johnson, R.N.; Roberts, C.W.; et al. Delivery of self-amplifying mRNA vaccines by cationic lipid nanoparticles: The impact of cationic lipid selection. J. Control. Release 2020, 325, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Kastner, E.; Kaur, R.; Lowry, D.; Moghaddam, B.; Wilkinson, A.; Perrie, Y. High-throughput manufacturing of size-tuned liposomes by a new microfluidics method using enhanced statistical tools for characterization. Int. J. Pharm. 2014, 477, 361–368. [Google Scholar] [CrossRef] [Green Version]
- Reichmuth, A.M.; Oberli, M.A.; Jeklenec, A.; Langer, R.; Blankschtein, D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016, 7, 319–334. [Google Scholar] [CrossRef] [Green Version]
- Tóth, E.L.; Holczer, E.G.; Iván, K.; Fürjes, P. Optimized simulation and validation of particle advection in asymmetric staggered herringbone type micromixers. Micromachines 2015, 6, 136–150. [Google Scholar] [CrossRef] [Green Version]
- Zizzari, A.; Bianco, M.; Carbone, L.; Perrone, E.; Amato, F.; Maruccio, G.; Rendina, F.; Arima, V. Continuous-flow production of injectable liposomes via a microfluidic approach. Materials 2017, 10, 1411. [Google Scholar] [CrossRef] [Green Version]
- Firmino, P.C.O.S.; Vianna, S.S.V.; da Costa, O.M.M.M.; Malfatti-Gasperini, A.A.; Gobbi, A.L.; Lima, R.S.; de la Torre, L.G. 3D micromixer for nanoliposome synthesis: A promising advance in high mass productivity. Lab Chip 2021, 21, 2971–2985. [Google Scholar] [CrossRef]
- Tripathi, E.; Patowari, P.K.; Pati, S. Numerical investigation of mixing performance in spiral micromixers based on Dean flows and chaotic advection. Chem. Eng. Process. Process Intensif. 2021, 169, 108609. [Google Scholar] [CrossRef]
- Xia, H.M.; Seah, Y.P.; Liu, Y.C.; Wang, W.; Toh, A.G.G.; Wang, Z.P. Anti-solvent precipitation of solid lipid nanoparticles using a microfluidic oscillator mixer. Microfluid. Nanofluidics 2015, 19, 283–290. [Google Scholar] [CrossRef]
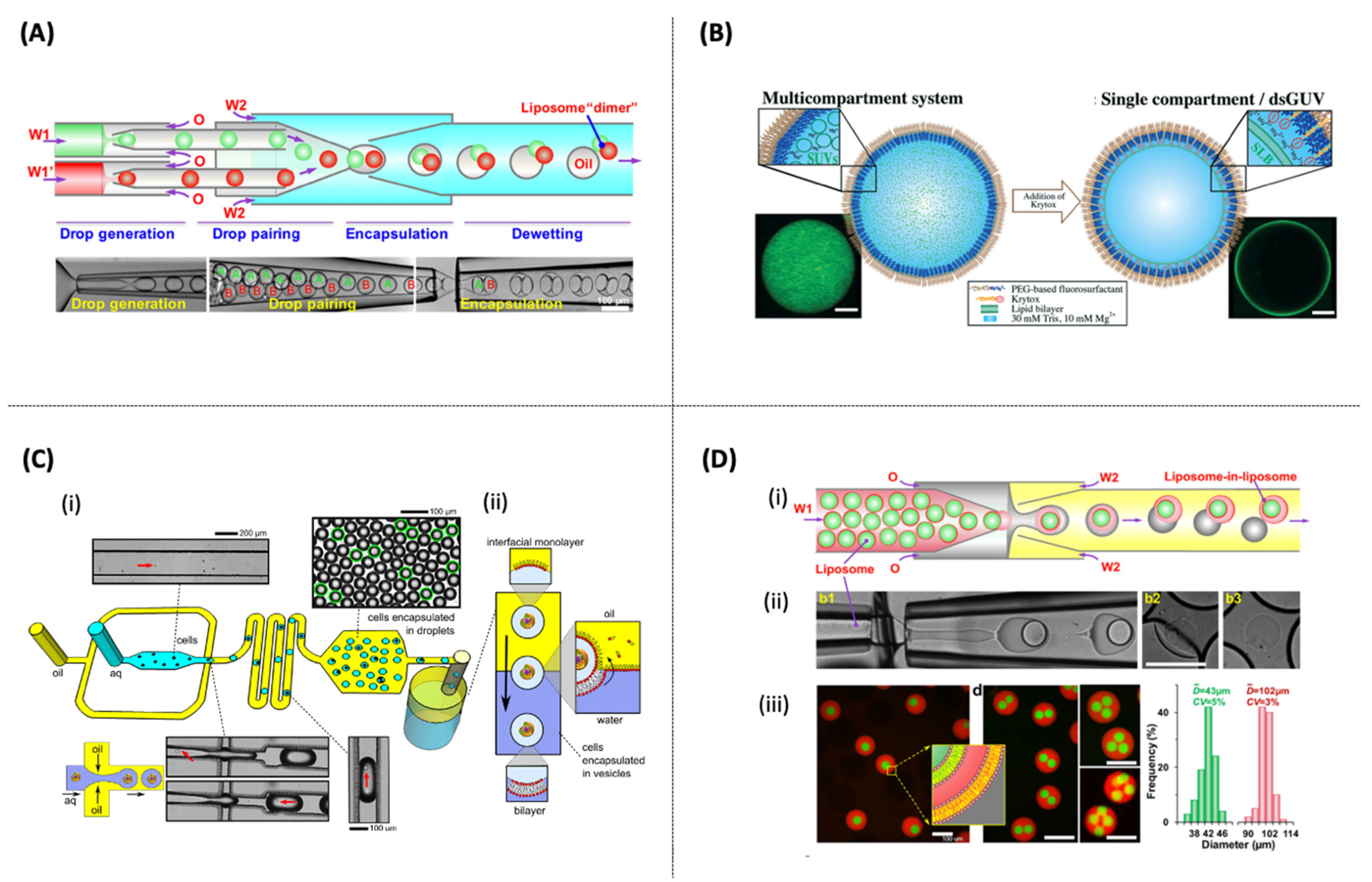
- Elani, Y.; Trantidou, T.; Wylie, D.; Dekker, L.; Polizzi, K.; Law, R.V.; Ces, O. Constructing vesicle-based artificial cells with embedded living cells as organelle-like modules. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, B.G.; Taketa, T.B.; Garcia, B.B.M.; Han, S.W.; de la Torre, L.G. Hybrid microgels produced via droplet microfluidics for sustainable delivery of hydrophobic and hydrophilic model nanocarriers. Mater. Sci. Eng. C 2021, 118, 111467. [Google Scholar] [CrossRef]
- Da Dong, Y.; Tchung, E.; Nowell, C.; Kaga, S.; Leong, N.; Mehta, D.; Kaminskas, L.M.; Boyd, B.J. Microfluidic preparation of drug-loaded PEGylated liposomes, and the impact of liposome size on tumour retention and penetration. J. Liposome Res. 2019, 29, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Hussain, M.T.; Roces, C.B.; Anderluzzi, G.; Kastner, E.; Salmaso, S.; Kirby, D.J.; Perrie, Y. Microfluidics based manufacture of liposomes simultaneously entrapping hydrophilic and lipophilic drugs. Int. J. Pharm. 2016, 514, 160–168. [Google Scholar] [CrossRef] [Green Version]
- Hamano, N.; Böttger, R.; Lee, S.E.; Yang, Y.; Kulkarni, J.A.; Ip, S.; Cullis, P.R.; Li, S.D. Robust microfluidic technology and new lipid composition for fabrication of curcumin-loaded liposomes: Effect on the anticancer activity and safety of cisplatin. Mol. Pharm. 2019, 16, 3957–3967. [Google Scholar] [CrossRef]
- Gkionis, L.; Campbell, R.A.; Aojula, H.; Harris, L.K.; Tirella, A. Manufacturing drug co-loaded liposomal formulations targeting breast cancer: Influence of preparative method on liposomes characteristics and in vitro toxicity. Int. J. Pharm. 2020, 590, 119926. [Google Scholar] [CrossRef] [PubMed]
- Ran, R.; Wang, H.; Liu, Y.; Hui, Y.; Sun, Q.; Seth, A.; Wibowo, D.; Chen, D.; Zhao, C.X. Microfluidic self-assembly of a combinatorial library of single- and dual-ligand liposomes for in vitro and in vivo tumor targeting. Eur. J. Pharm. Biopharm. 2018, 130, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balbino, T.A.; Serafin, J.M.; Radaic, A.; de Jesus, M.B.; de la Torre, L.G. Integrated microfluidic devices for the synthesis of nanoscale liposomes and lipoplexes. Colloids Surf. B Biointerfaces 2017, 152, 406–413. [Google Scholar] [CrossRef]
- Balbino, T.A.; Serafin, J.M.; Malfatti-Gasperini, A.A.; De Oliveira, C.L.P.; Cavalcanti, L.P.; De Jesus, M.B.; De La Torre, L.G. Microfluidic Assembly of pDNA/Cationic Liposome Lipoplexes with High pDNA Loading for Gene Delivery. Langmuir 2016. [Google Scholar] [CrossRef] [PubMed]
- Quagliarini, E.; Renzi, S.; Digiacomo, L.; Giulimondi, F.; Sartori, B.; Amenitsch, H.; Tassinari, V.; Masuelli, L.; Bei, R.; Cui, L.; et al. Microfluidic Formulation of DNA-Loaded Multicomponent Lipid Nanoparticles for Gene Delivery. Pharmaceutics 2021, 13, 1292. [Google Scholar] [CrossRef]
- Kimura, N.; Maeki, M.; Sato, Y.; Note, Y.; Ishida, A.; Tani, H.; Harashima, H.; Tokeshi, M. Development of the iLiNP Device: Fine Tuning the Lipid Nanoparticle Size within 10 nm for Drug Delivery. ACS Omega 2018, 3, 5044–5051. [Google Scholar] [CrossRef]
- Patel, S.; Ryals, R.C.; Weller, K.K.; Pennesi, M.E.; Sahay, G. Lipid nanoparticles for delivery of messenger RNA to the back of the eye. J. Control. Release 2019, 303, 91–100. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Myhre, J.L.; Chen, S.; Tam, Y.Y.C.; Danescu, A.; Richman, J.M.; Cullis, P.R. Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1377–1387. [Google Scholar] [CrossRef]
- Mucker, E.M.; Karmali, P.P.; Vega, J.; Kwilas, S.A.; Wu, H.; Joselyn, M.; Ballantyne, J.; Sampey, D.; Mukthavaram, R.; Sullivan, E.; et al. Lipid Nanoparticle Formulation Increases Efficiency of DNA-Vectored Vaccines/Immunoprophylaxis in Animals Including Transchromosomic Bovines. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Leung, J.; Van Der Meel, R.; Zaifman, J.; Darjuan, M.M.; Grisch-Chan, H.M.; Thöny, B.; Tam, Y.Y.C.; Cullis, P.R. Fusion-dependent formation of lipid nanoparticles containing macromolecular payloads. Nanoscale 2019, 11, 9023–9031. [Google Scholar] [CrossRef]
- Wei, W.; Sun, J.; Guo, X.Y.; Chen, X.; Wang, R.; Qiu, C.; Zhang, H.T.; Pang, W.H.; Wang, J.C.; Zhang, Q. Microfluidic-Based Holonomic Constraints of siRNA in the Kernel of Lipid/Polymer Hybrid Nanoassemblies for Improving Stable and Safe in Vivo Delivery. ACS Appl. Mater. Interfaces 2020, 12, 14839–14854. [Google Scholar] [CrossRef]
- Nie, T.; He, Z.; Zhou, Y.; Zhu, J.; Chen, K.; Liu, L.; Leong, K.W.; Mao, H.Q.; Chen, Y. Surface Coating Approach to Overcome Mucosal Entrapment of DNA Nanoparticles for Oral Gene Delivery of Glucagon-like Peptide 1. ACS Appl. Mater. Interfaces 2019, 11, 29593–29603. [Google Scholar] [CrossRef]
- Yang, R.; Deng, Y.; Huang, B.; Huang, L.; Lin, A.; Li, Y.; Wang, W.; Liu, J.; Lu, S.; Zhan, Z.; et al. A core-shell structured COVID-19 mRNA vaccine with favorable biodistribution pattern and promising immunity. Signal Transduct. Target. Ther. 2021, 6, 1–10. [Google Scholar] [CrossRef]
- Tahir, N.; Madni, A.; Li, W.; Correia, A.; Khan, M.M.; Rahim, M.A.; Santos, H.A. Microfluidic fabrication and characterization of Sorafenib-loaded lipid-polymer hybrid nanoparticles for controlled drug delivery. Int. J. Pharm. 2020, 581, 119275. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, J.; Wang, Y.; Wang, J.; Shi, X.; Hu, G. Nonspecific Organelle-Targeting Strategy with Core-Shell Nanoparticles of Varied Lipid Components/Ratios. Anal. Chem. 2016, 88, 7344–7351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, M.; Whittaker, A.K.; Jiang, X.; Tang, R.; Li, X.; Xu, W.; Fu, C.; Smith, M.T.; Han, F.Y. Use of Microfluidics to Fabricate Bioerodable Lipid Hybrid Nanoparticles Containing Hydromorphone or Ketamine for the Relief of Intractable Pain. Pharm. Res. 2020, 37, 1–12. [Google Scholar] [CrossRef]
- Bokare, A.; Takami, A.; Kim, J.H.; Dong, A.; Chen, A.; Valerio, R.; Gunn, S.; Erogbogbo, F. Herringbone-Patterned 3D-Printed Devices as Alternatives to Microfluidics for Reproducible Production of Lipid Polymer Hybrid Nanoparticles. ACS Omega 2019, 4, 4650–4657. [Google Scholar] [CrossRef] [Green Version]
- Molinaro, R.; Evangelopoulos, M.; Hoffman, J.R.; Corbo, C.; Taraballi, F.; Martinez, J.O.; Hartman, K.A.; Cosco, D.; Costa, G.; Romeo, I.; et al. Design and Development of Biomimetic Nanovesicles Using a Microfluidic Approach. Adv. Mater. 2018, 30, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, W.; Li, Y.; Chang, J.; Tian, F.; Zhao, F.; Ma, Y.; Sun, J. Microfluidic Sonication to Assemble Exosome Membrane-Coated Nanoparticles for Immune Evasion-Mediated Targeting. Nano Lett. 2019, 19, 7836–7844. [Google Scholar] [CrossRef]
- Zinger, A.; Cvetkovic, C.; Sushnitha, M.; Naoi, T.; Baudo, G.; Anderson, M.; Shetty, A.; Basu, N.; Covello, J.; Tasciotti, E.; et al. Humanized Biomimetic Nanovesicles for Neuron Targeting. Adv. Sci. 2021, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Kannisto, E.; Patnaik, S.K.; Reid, M.E.; Li, L.; Wu, Y. Ultrafast Detection of Exosomal RNAs via Cationic Lipoplex Nanoparticles in a Micromixer Biochip for Cancer Diagnosis. ACS Appl. Nano Mater. 2021, 4, 2806–2819. [Google Scholar] [CrossRef]
- Shah, S.; Dhawan, V.; Holm, R.; Nagarsenker, M.S.; Perrie, Y. Liposomes: Advancements and innovation in the manufacturing process. Adv. Drug Deliv. Rev. 2020, 154–155, 102–122. [Google Scholar] [CrossRef]
- Allen, T.M.; Cullis, P.R. Liposomal drug delivery systems: From concept to clinical applications. Adv. Drug Deliv. Rev. 2013, 65, 36–48. [Google Scholar] [CrossRef]
- Ilhan-ayisigi, E.; Ghazal, A.; Sartori, B.; Dimaki, M.; Svendsen, W.E.; Yesil-celiktas, O.; Yaghmur, A. Continuous microfluidic production of citrem-phosphatidylcholine nano-self-assemblies for thymoquinone delivery. Nanomaterials 2021, 11, 1510. [Google Scholar] [CrossRef]
- Costa, C.; Liu, Z.; Simões, S.I.; Correia, A.; Rahikkala, A.; Seitsonen, J.; Ruokolainen, J.; Aguiar-Ricardo, A.; Santos, H.A.; Corvo, M.L. One-step microfluidics production of enzyme-loaded liposomes for the treatment of inflammatory diseases. Colloids Surf. B Biointerfaces 2021, 199, 111556. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Szoka, F.C. Lipid-based nanoparticles for nucleic acid delivery. Pharm. Res. 2007, 24, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Eygeris, Y.; Patel, S.; Jozic, A.; Sahay, G. Deconvoluting Lipid Nanoparticle Structure for Messenger RNA Delivery. Nano Lett. 2020, 20, 4543–4549. [Google Scholar] [CrossRef]
- Leung, A.K.K.; Hafez, I.M.; Baoukina, S.; Belliveau, N.M.; Zhigaltsev, I.V.; Afshinmanesh, E.; Tieleman, D.P.; Hansen, C.L.; Hope, M.J.; Cullis, P.R. Lipid nanoparticles containing siRNA synthesized by microfluidic mixing exhibit an electron-dense nanostructured core. J. Phys. Chem. C 2012, 116, 18440–18450. [Google Scholar] [CrossRef]
- Samaridou, E.; Heyes, J.; Lutwyche, P. Lipid nanoparticles for nucleic acid delivery: Current perspectives. Adv. Drug Deliv. Rev. 2020, 154, 37–63. [Google Scholar] [CrossRef] [PubMed]
- Cullis, P.R.; Hope, M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017, 25, 1467–1475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef]
- Carvalho, B.G.; Vit, F.F.; Carvalho, H.F.; Han, S.W.; de la Torre, L.G. Recent advances in co-delivery nanosystems for synergistic action in cancer treatment. J. Mater. Chem. B 2020, 9, 1208–1237. [Google Scholar] [CrossRef]
- Maruggi, G.; Zhang, C.; Li, J.; Ulmer, J.B.; Yu, D. mRNA as a Transformative Technology for Vaccine Development to Control Infectious Diseases. Mol. Ther. 2019, 27, 757–772. [Google Scholar] [CrossRef] [Green Version]
- Dammes, N.; Peer, D. Paving the Road for RNA Therapeutics. Trends Pharmacol. Sci. 2020, 41, 755–775. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Karikó, K.; Türeci, Ö. mRNA-based therapeutics-developing a new class of drugs. Nat. Rev. Drug Discov. 2014, 13, 759–780. [Google Scholar] [CrossRef]
- Geall, A.J.; Verma, A.; Otten, G.R.; Shaw, C.A.; Hekele, A.; Banerjee, K.; Cu, Y.; Beard, C.W.; Brito, L.A.; Krucker, T.; et al. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA 2012, 109, 14604–14609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bloom, K.; van den Berg, F.; Arbuthnot, P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 2021, 28, 117–129. [Google Scholar] [CrossRef]
- Pushko, P.; Parker, M.; Ludwig, G.V.; Davis, N.L.; Johnston, R.E.; Smith, J.F. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: Expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology 1997, 239, 389–401. [Google Scholar] [CrossRef] [Green Version]
- Whitehead, K.A.; Langer, R.; Anderson, D.G. Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov. 2009, 8, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Dykxhoorn, D.M.; Lieberman, J. Running interference: Prospects and obstacles to using small interfering RNAs as small molecule drugs. Annu. Rev. Biomed. Eng. 2006, 8, 377–402. [Google Scholar] [CrossRef] [Green Version]
- Akinc, A.; Maier, M.A.; Manoharan, M.; Fitzgerald, K.; Jayaraman, M.; Barros, S.; Ansell, S.; Du, X.; Hope, M.J.; Madden, T.D.; et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019, 14, 1084–1087. [Google Scholar] [CrossRef]
- Chen, S.; Tam, Y.Y.C.; Lin, P.J.C.; Sung, M.M.H.; Tam, Y.K.; Cullis, P.R. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Control. Release 2016, 235, 236–244. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Lin, P.J.C.; Beraldi, E.; Zhang, F.; Kawai, Y.; Leong, J.; Katsumi, H.; Fazli, L.; Fraser, R.; Cullis, P.R.; et al. siRNA lipid nanoparticle potently silences clusterin and delays progression when combined with androgen receptor cotargeting in enzalutamide-resistant prostate cancer. Clin. Cancer Res. 2015, 21, 4845–4855. [Google Scholar] [CrossRef] [Green Version]
- Yanagi, T.; Tachikawa, K.; Wilkie-Grantham, R.; Hishiki, A.; Nagai, K.; Toyonaga, E.; Chivukula, P.; Matsuzawa, S. ichi Lipid Nanoparticle-mediated siRNA Transfer Against PCTAIRE1/PCTK1/Cdk16 Inhibits In Vivo Cancer Growth. Mol. Ther. Nucleic Acids 2016, 5, 327. [Google Scholar] [CrossRef] [Green Version]
- Jyotsana, N.; Sharma, A.; Chaturvedi, A.; Budida, R.; Scherr, M.; Kuchenbauer, F.; Lindner, R.; Noyan, F.; Sühs, K.-W.; Stangel, M.; et al. Lipid nanoparticle-mediated siRNA delivery for safe targeting of human CML in vivo. Ann. Hematol. 2019, 98, 1905–1918. [Google Scholar] [CrossRef]
- Billingsley, M.M.; Singh, N.; Ravikumar, P.; Zhang, R.; June, C.H.; Mitchell, M.J. Ionizable Lipid Nanoparticle-Mediated mRNA Delivery for Human CAR T Cell Engineering. Nano Lett. 2020, 20, 1578–1589. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.S.; Kashyap, M.V.; Billingsley, M.M.; White, B.; Alameh, M.G.; Bose, S.K.; Zoltick, P.W.; Li, H.; Zhang, R.; Cheng, A.Y.; et al. Ionizable lipid nanoparticles for in utero mRNA delivery. Sci. Adv. 2021, 7, eaba1028. [Google Scholar] [CrossRef] [PubMed]
- Mandal, B.; Bhattacharjee, H.; Mittal, N.; Sah, H.; Balabathula, P.; Thoma, L.A.; Wood, G.C. Core-shell-type lipid-polymer hybrid nanoparticles as a drug delivery platform. Nanomed. Nanotechnol. Biol. Med. 2013, 9, 474–491. [Google Scholar] [CrossRef]
- Date, T.; Nimbalkar, V.; Kamat, J.; Mittal, A.; Mahato, R.I.; Chitkara, D. Lipid-polymer hybrid nanocarriers for delivering cancer therapeutics. J. Control. Release 2018, 271, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Hadinoto, K.; Sundaresan, A.; Cheow, W.S. Lipid-polymer hybrid nanoparticles as a new generation therapeutic delivery platform: A review. Eur. J. Pharm. Biopharm. 2013, 85, 427–443. [Google Scholar] [CrossRef]
- Hu, H.; Yang, C.; Li, M.; Shao, D.; Mao, H.Q.; Leong, K.W. Flash technology-based self-assembly in nanoformulation: Fabrication to biomedical applications. Mater. Today 2021, 42, 99–116. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Kang, M.-H.; Jeyaraj, M.; Qasim, M.; Kim, J.-H. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Discher, D.; Eisenberg, A. Polymer Vesicles. Science 2002, 297, 967–973. [Google Scholar] [CrossRef] [Green Version]
- Yandrapalli, N.; Petit, J.; Bäumchen, O.; Robinson, T. Surfactant-free production of biomimetic giant unilamellar vesicles using PDMS-based microfluidics. Commun. Chem. 2021, 4, 1–10. [Google Scholar] [CrossRef]
- Rigaud, J.L.; Pitard, B.; Levy, D. Reconstitution of membrane proteins into liposomes: Application to energy-transducing membrane proteins. BBA Bioenergy 1995, 1231, 223–246. [Google Scholar] [CrossRef] [Green Version]
- Aimon, S.; Manzi, J.; Schmidt, D.; Larrosa, J.A.P.; Bassereau, P.; Toombes, G.E.S. Functional reconstitution of a voltage-gated potassium channel in giant unilamellar vesicles. PLoS ONE 2011, 6, e25529. [Google Scholar] [CrossRef]
- Girard, P.; Pécréaux, J.; Lenoir, G.; Falson, P.; Rigaud, J.L.; Bassereau, P. A new method for the reconstitution of membrane proteins into giant unilamellar vesicles. Biophys. J. 2004, 87, 419–429. [Google Scholar] [CrossRef] [Green Version]
- Montes, L.R.; Alonso, A.; Goñi, F.M.; Bagatolli, L.A. Giant unilamellar vesicles electroformed from native membranes and organic lipid mixtures under physiological conditions. Biophys. J. 2007, 93, 3548–3554. [Google Scholar] [CrossRef] [Green Version]
- Stein, H.; Spindler, S.; Bonakdar, N.; Wang, C.; Sandoghdar, V. Production of isolated giant unilamellar vesicles under high salt concentrations. Front. Physiol. 2017, 8, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Pereno, V.; Carugo, D.; Bau, L.; Sezgin, E.; De La Serna, J.B.; Eggeling, C.; Stride, E. Electroformation of Giant Unilamellar Vesicles on Stainless Steel Electrodes. ACS Omega 2017, 2, 994–1002. [Google Scholar] [CrossRef] [Green Version]
- Horger, K.S.; Estes, D.J.; Capone, R.; Mayer, M. Films of agarose enable rapid formation of giant liposomes in solutions of physiologic ionic strength. J. Am. Chem. Soc. 2009, 131, 1810–1819. [Google Scholar] [CrossRef] [Green Version]
- Pautot, S.; Frisken, B.J.; Weitz, D.A. Engineering asymmetric vesicles. Proc. Natl. Acad. Sci. USA 2003, 100, 10718–10721. [Google Scholar] [CrossRef] [Green Version]
- Elani, Y. Construction of membrane-bound artificial cells using microfluidics: A new frontier in bottom-up synthetic biology. Biochem. Soc. Trans. 2016, 44, 723–730. [Google Scholar] [CrossRef]
- Seo, H.; Lee, H. Recent developments in microfluidic synthesis of artificial cell-like polymersomes and liposomes for functional bioreactors. Biomicrofluidics 2021, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Weiss, M.; Frohnmayer, J.P.; Benk, L.T.; Haller, B.; Janiesch, J.-W.; Heitkamp, T.; Börsch, M.; Lira, R.B.; Dimova, R.; Lipowsky, R.; et al. Sequential bottom-up assembly of mechanically stabilized synthetic cells by microfluidics. Nat. Mater. 2018, 17, 89–96. [Google Scholar] [CrossRef]
- Petit, J.; Polenz, I.; Baret, J.C.; Herminghaus, S.; Bäumchen, O. Vesicles-on-a-chip: A universal microfluidic platform for the assembly of liposomes and polymersomes. Eur. Phys. J. E 2016, 39, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaich, M.; Cama, J.; Al Nahas, K.; Sobota, D.; Sleath, H.; Jahnke, K.; Deshpande, S.; Dekker, C.; Keyser, U.F. An Integrated Microfluidic Platform for Quantifying Drug Permeation across Biomimetic Vesicle Membranes. Mol. Pharm. 2019, 16, 2494–2501. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, S.; Dekker, C. On-chip microfluidic production of cell-sized liposomes. Nat. Protoc. 2018, 13, 856–874. [Google Scholar] [CrossRef]
- Shum, H.C.; Lee, D.; Yoon, I.; Kodger, T.; Weitz, D.A. Double emulsion templated monodisperse phospholipid vesicles. Langmuir 2008, 24, 7651–7653. [Google Scholar] [CrossRef] [PubMed]
- Arriaga, L.R.; Huang, Y.; Kim, S.H.; Aragones, J.L.; Ziblat, R.; Koehler, S.A.; Weitz, D.A. Single-step assembly of asymmetric vesicles. Lab Chip 2019, 19, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Van Swaay, D.; Demello, A. Microfluidic methods for forming liposomes. Lab Chip 2013, 13, 752–767. [Google Scholar] [CrossRef] [PubMed]
- Deng, N.N.; Yelleswarapu, M.; Huck, W.T.S. Monodisperse Uni- and Multicompartment Liposomes. J. Am. Chem. Soc. 2016, 138, 7584–7591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haller, B.; Göpfrich, K.; Schröter, M.; Janiesch, J.W.; Platzman, I.; Spatz, J.P. Charge-controlled microfluidic formation of lipid-based single- and multicompartment systems. Lab Chip 2018, 18, 2665–2674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.C.; Hettiarachchi, K.; Siu, M.; Pan, Y.R.; Lee, A.P. Controlled microfluidic encapsulation of cells, proteins, and microbeads in lipid vesicles. J. Am. Chem. Soc. 2006, 128, 5656–5658. [Google Scholar] [CrossRef]
- Deng, N.N.; Yelleswarapu, M.; Zheng, L.; Huck, W.T.S. Microfluidic assembly of monodisperse vesosomes as artificial cell models. J. Am. Chem. Soc. 2017, 139, 587–590. [Google Scholar] [CrossRef] [Green Version]
- Deshpande, S.; Caspi, Y.; Meijering, A.E.C.; Dekker, C. Octanol-assisted liposome assembly on chip. Nat. Commun. 2016, 7, 1–9. [Google Scholar] [CrossRef]
- Czekalska, M.A.; Jacobs, A.M.J.; Toprakcioglu, Z.; Kong, L.; Baumann, K.N.; Gang, H.; Zubaite, G.; Ye, R.; Mu, B.; Levin, A.; et al. One-Step Generation of Multisomes from Lipid-Stabilized Double Emulsions. ACS Appl. Mater. Interfaces 2021, 13, 6739–6747. [Google Scholar] [CrossRef]
- Kamiya, K.; Takeuchi, S. Giant liposome formation toward the synthesis of well-defined artificial cells. J. Mater. Chem. B 2017, 5, 5911–5923. [Google Scholar] [CrossRef]
- Hoare, T.R.; Kohane, D.S. Hydrogels in drug delivery: Progress and challenges. Polymer 2008, 49, 1993–2007. [Google Scholar] [CrossRef] [Green Version]
- Liechty, W.B.; Kryscio, D.R.; Slaughter, B.V.; Peppas, N.A. Polymers for Drug Delivery Systems. Annu. Rev. Chem. Biomol. Eng. 2010, 1, 149–173. [Google Scholar] [CrossRef] [Green Version]
- Takagi, I.; Shimizu, H.; Yotsuyanagi, T. Application of alginate gel as a vehicle for liposomes. I. Factors affecting the loading of drug-containing liposomes and drug release. Chem. Pharm. Bull. 1996, 44, 1941–1947. [Google Scholar] [CrossRef] [Green Version]
- Grijalvo, S.; Mayr, J.; Eritja, R.; Díaz, D.D. Biodegradable liposome-encapsulated hydrogels for biomedical applications: A marriage of convenience. Biomater. Sci. 2016, 4, 555–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulkarni, M.; Breen, A.; Greiser, U.; O’Brien, T.; Pandit, A. Fibrin-lipoplex system for controlled topical delivery of multiple genes. Biomacromolecules 2009, 10, 1650–1654. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, M.M.; Greiser, U.; O’Brien, T.; Pandit, A. A temporal gene delivery system based on fibrin microspheres. Mol. Pharm. 2011, 8, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Sacks, H.; Elazar, V.; Gao, J.; Golomb, A.; Adwan, H.; Korchov, N.; Levy, R.J.; Berger, M.R.; Golomb, G. Delivery and expression of pDNA embedded in collagen matrices. J. Control. Release 2004, 95, 309–320. [Google Scholar] [CrossRef]
- Wu, S.Y.; Chang, H.I.; Burgess, M.; McMillan, N.A.J. Vaginal delivery of siRNA using a novel PEGylated lipoplex-entrapped alginate scaffold system. J. Control. Release 2011, 155, 418–426. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Zhang, H.; Yan, J.; Bryers, J.D. Scaffold-mediated delivery for non-viral mRNA vaccines. Gene Ther. 2018, 25, 556–567. [Google Scholar] [CrossRef]
- Daly, A.C.; Riley, L.; Segura, T.; Burdick, J.A. Hydrogel microparticles for biomedical applications. Nat. Rev. Mater. 2020, 5, 20–43. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.T.; Wang, J.; Han, J.J. Fabrication of advanced particles and particle-based materials assisted by droplet-based microfluidics. Small 2011, 7, 1728–1754. [Google Scholar] [CrossRef]
- Thiele, J. Polymer Material Design by Microfluidics Inspired by Cell Biology and Cell-Free Biotechnology. Macromol. Chem. Phys. 2017, 218, 1–16. [Google Scholar] [CrossRef]
- Windbergs, M.; Zhao, Y.; Heyman, J.; Weitz, D.A. Biodegradable Core − Shell Carriers for Simultaneous Encapsulation of Synergistic Actives. J. Am. Chem. Soc. 2013, 135, 7933–7937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazutis, L.; Vasiliauskas, R.; Weitz, D.A. Microfluidic Production of Alginate Hydrogel Particles for Antibody Encapsulation and Release. Macromol. Biosci. 2015, 15, 1641–1646. [Google Scholar] [CrossRef]
- Deveza, L.; Ashoken, J.; Castaneda, G.; Tong, X.; Keeney, M.; Han, L.H.; Yang, F. Microfluidic Synthesis of Biodegradable Polyethylene-Glycol Microspheres for Controlled Delivery of Proteins and DNA Nanoparticles. ACS Biomater. Sci. Eng. 2015, 1, 157–165. [Google Scholar] [CrossRef]
- Cinel, V.D.P.; Taketa, T.B.; Carvalho, B.G.; de la Torre, L.G.; de Mello, L.R.; da Silva, E.R.; Han, S.W. Microfluidic encapsulation of nanoparticles in alginate microgels gelled via competitive ligand exchange crosslinking. Biopolymers 2021, 112, 23432. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.; Liu, Z.; Martins, J.P.; Correia, A.; Figueiredo, P.; Rahikkala, A.; Li, W.; Seitsonen, J.; Ruokolainen, J.; Hirvonen, S.P.; et al. All-in-one microfluidic assembly of insulin-loaded pH-responsive nano-in-microparticles for oral insulin delivery. Biomater. Sci. 2020, 8, 3270–3277. [Google Scholar] [CrossRef] [PubMed]
- Jeong, E.S.; Son, H.A.; Kim, M.K.; Park, K.H.; Kay, S.; Chae, P.S.; Kim, J.W. Fabrication of monodisperse liposomes-in-microgel hybrid microparticles in capillary-based microfluidic devices. Colloids Surf. B Biointerfaces 2014, 123, 339–344. [Google Scholar] [CrossRef]
- Pittermannová, A.; Ruberová, Z.; Zadražil, A.; Bremond, N.; Bibette, J.; Štěpánek, F. Microfluidic fabrication of composite hydrogel microparticles in the size range of blood cells. RSC Adv. 2016, 6, 103532–103540. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhu, Y.; Wang, F.; Deng, L.; Xu, X.; Cui, W. Microfluidic liposomes-anchored microgels as extended delivery platform for treatment of osteoarthritis. Chem. Eng. J. 2020, 400, 126004. [Google Scholar] [CrossRef]
- Madrigal, J.L.; Stilhano, R.S.; Siltanen, C.; Tanaka, K.; Rezvani, S.N.; Morgan, R.P.; Revzin, A.; Han, S.W.; Silva, E.A. Microfluidic generation of alginate microgels for the controlled delivery of lentivectors. J. Mater. Chem. B 2016, 4, 6989–6999. [Google Scholar] [CrossRef] [Green Version]
- Madrigal, J.L.; Sharma, S.N.; Campbell, K.T.; Stilhano, R.S.; Gijsbers, R.; Silva, E.A. Microgels produced using microfluidic on-chip polymer blending for controlled released of VEGF encoding lentivectors. Acta Biomater. 2018, 69, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, B.G.; Vit, F.F.; Carvalho, H.F.; Han, S.W.; Torre, L.G. De Layered Biomimetic Microgels for 3D Cell Culture and Nonviral Gene Delivery. Biomacromolecules 2021, 15, 1003–1004. [Google Scholar] [CrossRef]
- Seo, M.; Byun, A.; Shim, J.; Choi, H.S.; Lee, Y.; Kim, J.W. Uniform and stable hydrogel-filled liposome-analogous vesicles with a thin elastomer shell layer. Colloids Surf. B Biointerfaces 2016, 146, 544–549. [Google Scholar] [CrossRef]
- Wang, H.F.; Ran, R.; Liu, Y.; Hui, Y.; Zeng, B.; Chen, D.; Weitz, D.A.; Zhao, C.X. Tumor-Vasculature-on-a-Chip for Investigating Nanoparticle Extravasation and Tumor Accumulation. ACS Nano 2018, 12, 11600–11609. [Google Scholar] [CrossRef]
- Li, X.; Aghaamoo, M.; Liu, S.; Lee, D.H.; Lee, A.P. Lipoplex-Mediated Single-Cell Transfection via Droplet Microfluidics. Small 2018, 14, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Kwak, B.; Ozcelikkale, A.; Shin, C.S.; Park, K.; Han, B. Simulation of complex transport of nanoparticles around a tumor using tumor-microenvironment-on-chip. J. Control. Release 2014, 194, 157–167. [Google Scholar] [CrossRef] [Green Version]
- Ozcelikkale, A.; Shin, K.; Noe-Kim, V.; Elzey, B.D.; Dong, Z.; Zhang, J.T.; Kim, K.; Kwon, I.C.; Park, K.; Han, B. Differential response to doxorubicin in breast cancer subtypes simulated by a microfluidic tumor model. J. Control. Release 2017, 266, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Park, S.E.; Huh, D.D. Organ-on-a-chip technology for nanoparticle research. Nano Converg. 2021, 8, 1–15. [Google Scholar] [CrossRef]
- Papademetriou, I.; Vedula, E.; Charest, J.; Porter, T. Effect of flow on targeting and penetration of angiopep-decorated nanoparticles in a microfluidic model blood-brain barrier. PLoS ONE 2018, 13, e0205158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ran, R.; Wang, H.F.; Hou, F.; Liu, Y.; Hui, Y.; Petrovsky, N.; Zhang, F.; Zhao, C.X. A Microfluidic Tumor-on-a-Chip for Assessing Multifunctional Liposomes’ Tumor Targeting and Anticancer Efficacy. Adv. Healthc. Mater. 2019, 8, 1–10. [Google Scholar] [CrossRef]
- Tang, Y.; Soroush, F.; Sheffield, J.B.; Wang, B.; Prabhakarpandian, B.; Kiani, M.F. A Biomimetic Microfluidic Tumor Microenvironment Platform Mimicking the EPR Effect for Rapid Screening of Drug Delivery Systems. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Paek, J.; Park, S.E.; Lu, Q.; Park, K.T.; Cho, M.; Oh, J.M.; Kwon, K.W.; Yi, Y.S.; Song, J.W.; Edelstein, H.I.; et al. Microphysiological Engineering of Self-Assembled and Perfusable Microvascular Beds for the Production of Vascularized Three-Dimensional Human Microtissues. ACS Nano 2019, 13, 7627–7643. [Google Scholar] [CrossRef]
- Virumbrales-Muñoz, M.; Ayuso, J.M.; Olave, M.; Monge, R.; De Miguel, D.; Martínez-Lostao, L.; Le Gac, S.; Doblare, M.; Ochoa, I.; Fernandez, L.J. Multiwell capillarity-based microfluidic device for the study of 3D tumour tissue-2D endothelium interactions and drug screening in co-culture models. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Li, J.; Wen, A.M.; Potla, R.; Benshirim, E.; Seebarran, A.; Benz, M.A.; Henry, O.Y.F.; Matthews, B.D.; Prantil-Baun, R.; Gilpin, S.E.; et al. AAV-mediated gene therapy targeting TRPV4 mechanotransduction for inhibition of pulmonary vascular leakage. APL Bioeng. 2019, 3, 046103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Kim, S.; Koo, D.J.; Yu, J.; Cho, H.; Lee, H.; Song, J.M.; Kim, S.Y.; Min, D.H.; Jeon, N.L. 3D Microfluidic Platform and Tumor Vascular Mapping for Evaluating Anti-Angiogenic RNAi-Based Nanomedicine. ACS Nano 2021, 15, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Vitor, M.T.; Sart, S.; Barizien, A.; La Torre, L.G.D.; Baroud, C.N. Tracking the Evolution of Transiently Transfected Individual Cells in a Microfluidic Platform. Sci. Rep. 2018, 8, 25–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raimes, W.; Rubi, M.; Super, A.; Marques, M.P.C.; Veraitch, F.; Szita, N. Transfection in perfused microfluidic cell culture devices: A case study. Process Biochem. 2017, 59, 297–302. [Google Scholar] [CrossRef] [Green Version]
- Sharei, A.; Zoldan, J.; Adamo, A.; Sim, W.Y.; Cho, N.; Jackson, E.; Mao, S.; Schneider, S.; Han, M.J.; Lytton-Jean, A.; et al. A vector-free microfluidic platform for intracellular delivery. Proc. Natl. Acad. Sci. USA 2013, 110, 2082–2087. [Google Scholar] [CrossRef] [Green Version]
- Szeto, G.L.; Van Egeren, D.; Worku, H.; Sharei, A.; Alejandro, B.; Park, C.; Frew, K.; Brefo, M.; Mao, S.; Heimann, M.; et al. Microfluidic squeezing for intracellular antigen loading in polyclonal B-cells as cellular vaccines. Sci. Rep. 2015, 5, 1–13. [Google Scholar] [CrossRef]
- Tran, R.; Myers, D.R.; Denning, G.; Shields, J.E.; Lytle, A.M.; Alrowais, H.; Qiu, Y.; Sakurai, Y.; Li, W.C.; Brand, O.; et al. Microfluidic Transduction Harnesses Mass Transport Principles to Enhance Gene Transfer Efficiency. Mol. Ther. 2017, 25, 2372–2382. [Google Scholar] [CrossRef] [Green Version]
- Jarrell, J.A.; Twite, A.A.; Lau, K.H.W.J.; Kashani, M.N.; Lievano, A.A.; Acevedo, J.; Priest, C.; Nieva, J.; Gottlieb, D.; Pawell, R.S. Intracellular delivery of mRNA to human primary T cells with microfluidic vortex shedding. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giupponi, E.; Visone, R.; Occhetta, P.; Colombo, F.; Rasponi, M.; Candiani, G. Development of a microfluidic platform for high-throughput screening of non-viral gene delivery vectors. Biotechnol. Bioeng. 2018, 115, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Probst, J.; Borca, C.N.; Newton, M.A.; van Bokhoven, J.; Huthwelker, T.; Stavrakis, S.; de Mello, A. In Situ X-ray Absorption Spectroscopy and Droplet-Based Microfluidics: An Analysis of Calcium Carbonate Precipitation. ACS Meas. Sci. Au 2021, 1, 27–34. [Google Scholar] [CrossRef]
- Poulos, A.S.; Nania, M.; Lapham, P.; Miller, R.M.; Smith, A.J.; Tantawy, H.; Caragay, J.; Gummel, J.; Ces, O.; Robles, E.S.J.; et al. Microfluidic SAXS Study of Lamellar and Multilamellar Vesicle Phases of Linear Sodium Alkylbenzenesulfonate Surfactant with Intrinsic Isomeric Distribution. Langmuir 2016, 32, 5852–5861. [Google Scholar] [CrossRef] [Green Version]
- Adamo, M.; Poulos, A.S.; Lopez, C.G.; Martel, A.; Porcar, L.; Cabral, J.T. Droplet microfluidic SANS. Soft Matter 2018, 14, 1759–1770. [Google Scholar] [CrossRef]
- Liu, L.; Bi, M.; Wang, Y.; Liu, J.; Jiang, X.; Xu, Z.; Zhang, X. Artificial intelligence-powered microfluidics for nanomedicine and materials synthesis. Nanoscale 2021, 13, 19352–19366. [Google Scholar] [CrossRef]



| Type of Nanocarrier | Nanocarrier Composition | Therapeutics | Microfluidic Device Type | Potential Application | Ref. |
|---|---|---|---|---|---|
| Liposome | PC and CHOL | Propofol | SHM * | Anesthesia | [68] |
| PC, DMPC, DPPC, CHOL, PS, and DSPC | Insulin, BSA, or OVA | SHM * | - | [69] | |
| HSPC, CHOL, and DSPE-PEG2000 | DOX | N/A * | Cancer: MDA-MB231cells and xenograft model bearing MDA-MB231tumor | [82] | |
| PC, DMPC, DPPC, and DSPC | Glipizide and metformin | SHM * | Diabetes | [83] | |
| DMPC, DPPC, and DSPC | Cisplatin and Curcumin | SHM * | Cancer: EMT6 and B16F10 cells/and xenograft model bearing EMT6 and B16F10 tumor | [84] | |
| DSPC, CHOL, and DSPE-PEG2000 | DOX and UMB | 5-Input Chip ** | Cancer: MCF-7, MDA-MB231, and BT-473 cells | [85] | |
| DMPC, DSPE-PEG, and CHOL. Ligands: DSPE-PEG-TAT and DSPE-PEG-Folate | - | HFF device | Cancer: SKOV3 and MCF-7 cells and 3D tumor spheroids/and xenograft model bearing SKOV3 tumor | [86] | |
| Lipoplexes | cationic lipid DOTAP, EPC, and DOPE | pEGFP-N1 | HFF device | PC3 cells | [87,88] |
| cationic lipid (DOTAP, DDA, DC-CHOL, DMTAP, DSTAP or DOBAQ), DOPE or DSPC, CHOL, and DMG-PEG2000 | SAM encoding rabies virus glycoprotein (RVG) | Y-shape SHM * | Prophylactic vaccine:BHK cells and BALB/c mice | [72] | |
| cationic lipid DOTAP, DC-CHOL, DOPE, CHOL, and DOPE-PEG | pGL3 | Y-shape SHM * | HEK-293, HaCaT, N/TERT, and CaSki cells | [89] | |
| pH-sensitive cationic lipid YSK05, chol, and PEG-DMG | siFVII | Baffle mixer device | ICR mice liver tissues: hepatocytes delivery and FVII gene-silencing activity. | [90] | |
| iLNP and cLNP | Ionizable lipid MC3 or cationic lipids DOTAP and DDAB, HSPC or DSPC, CHOL, and DMG-PEG2000 or DSPE-PEG2000 | PolyA, ssDNA or mFLuc | Y-shape SHM * | N/A | [70] |
| Ionizable lipid (MC3, KC2, DODMA) or cationic lipid (DOBAQ, DOTMA, DOTAP), DSPC, DMG-PEG2k, and CHOL | mRNAs: mFLuc, mEGFP, and mCherry | N/A | Retinal degeneration: BALB/c mice | [91] | |
| iLNP | Ionizable lipid (C12−200), DOPE or DSPC, CHOL, and lipid-PEG | siFVII or mLuc | SHM parallelized device | HeLa cells and C57BL/6 mice | [21] |
| Ionizable lipids MC3 or KC2, DLinDAP or DLinDMA, CHOL, DOPE, DOPC, SOPC, DLinPC, DPoPC or DSPC, and DMG-PEG | pDNAs: pEGFP or pFLuc | T-junction mixer | HeLa, HepG2, Hep3B, PC12, and MCF7 cells (in vitro) and leghorn chicken embryos (in vivo) | [92] | |
| ATX ionizable amino lipids, CHOL, DSPC, and DMG-PEG | pWRG/c7d11 | N/A* | Prophylactic Andes and Zika virus vaccine: Vero cells, rabbits, and nonhuman primates | [93] | |
| Ionizable lipid KC2, CHOL, DSPC, and DMG-PEG | mFLuc or mcDNA | T-junction mixer | N/A | [94] |
| Type of Nanocarrier | Nanocarrier Composition | Therapeutics | Microfluidic Device Type | Potential Application | Ref. |
|---|---|---|---|---|---|
| CORE: PCL-PEI/SHELL: CHOL, DSPE-PEG2000, and DOPE | siEGFR | Three-stage microfluidic chip (MiTASChip Ltd., Jiangsu, China) | Cancer: PC3 cells and xenograft model bearing PC-3 tumor | [95] | |
| CORE: PEI/SHELL: CHOL, DPPC, and DMG-PEG | pGLP-1 | CIJM and MIVM | Oral delivery type II diabetes-293T, A549, HepG2, HeLa cells, and BALB/c mice | [96] | |
| CORE: Cationic material (SW-01)/SHELL: ionizable lipid, DOPE, and PEG-lipid | mRNAs: mEGFP and mSARS-CoV-2 Spike (S) (in vitro)/mLuc (in vivo) | Two-step microfluidic mixer (Inano D, Micro&Nano Technology Inc., China) | Prophylactic COVID vaccine: DC 2.4, HEK-293 T cells, and BALB/c mice | [97] | |
| CORE: PLGA/SHELL: Lecithin and DSPE-PEG 2000 | Sorafenib | Borosilicate glass capillaries | Cancer: MDA-MB231, PC3-MM2, and HT29-MTX cells | [98] | |
| CORE: PLGA/SHELL: DOTAP, DOPE, CHOL, DPPC, and DSPE-PEG | - | Two-stage microfluidic device | HUVEC cells and BALB/c mice | [99] | |
| CORE: PLGA and CPP-SA/SHELL: DPPC, DSPE-PEG, and CHOL | Ketamine and hydromorphone | Two-stage microfluidic device | Intractable neuropathic pain: Chronic constriction injury (CCI)-rats | [100] | |
| CORE: PLGA/SHELL: Lecithin and DSPE-PEG | Rifampicin | MIVM and herringbone-patterned MIVM | Tuberculosis | [101] | |
| BIOMIMETIC VESICLES | LIPIDS: DPPC, DOPC and CHOL/PROTEIN: leukocyte membrane proteins | - | N/A * | J774 macrophages | [102] |
| CORE: PLGA/SHELL: cancer cell or exosome membranes or lipids (DPPC, CHOL, and DSPE-PEG) | - | Two-stage microfluidic device | Cancer: A549, MDA-MB-231, RAW 264.7 cells, and xenograft model bearing A549 and MDA-MB-231tumors | [103] | |
| LIPIDS: (1) DPPC, DOPC, and CHOL and (2) DAP, DSPE-PEG2000, and CHOL/PROTEIN: hPSC-derived excitatory cortical neurons | - | N/A * | Human pluripotent stem cells (hPSCs) and trigeminal ganglion of C57BL/6mice | [104] | |
| LIPIDS: DOTMA, CHOL, TPGS | Molecular beacons: TPGS exosomal RNA FAM-miR-21 MBs and Cy5-TTF-1 MBs | Layer-by-layer micromixer biochip | Cancer: A549 NSCLC and BEAS-2B cells | [105] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carvalho, B.G.; Ceccato, B.T.; Michelon, M.; Han, S.W.; de la Torre, L.G. Advanced Microfluidic Technologies for Lipid Nano-Microsystems from Synthesis to Biological Application. Pharmaceutics 2022, 14, 141. https://doi.org/10.3390/pharmaceutics14010141
Carvalho BG, Ceccato BT, Michelon M, Han SW, de la Torre LG. Advanced Microfluidic Technologies for Lipid Nano-Microsystems from Synthesis to Biological Application. Pharmaceutics. 2022; 14(1):141. https://doi.org/10.3390/pharmaceutics14010141
Chicago/Turabian StyleCarvalho, Bruna G., Bruno T. Ceccato, Mariano Michelon, Sang W. Han, and Lucimara G. de la Torre. 2022. "Advanced Microfluidic Technologies for Lipid Nano-Microsystems from Synthesis to Biological Application" Pharmaceutics 14, no. 1: 141. https://doi.org/10.3390/pharmaceutics14010141







