Exploiting Pharma 4.0 Technologies in the Non-Biological Complex Drugs Manufacturing: Innovations and Implications
Abstract
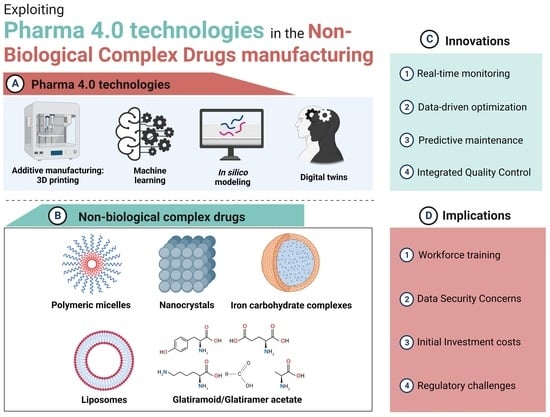
:1. Introduction
2. Emerging Tools of Pharma 4.0
2.1. Additive Manufacturing: Three-Dimensional (3D) Printing
Advancements in the Pharmaceutical Industry Offered by 3D Printing Technology
2.2. In Silico Modeling
2.3. Machine Learning (ML)
2.4. Digital Twins
3. Non-Biological Complex Drugs (NBCDs)
3.1. Polymeric Micelles
3.2. Liposomes
3.2.1. Niosomes
3.2.2. Transfersomes
3.3. Glatiramoid/Glatiramer Acetate (GA)
3.4. Iron Carbohydrate Complexes Drugs
3.5. Nanocrystals
4. Applying Pharma 4.0 Tools in the Production of Non-Biological Complex Drugs
4.1. Additive Manufacturing: 3D Printing
4.1.1. Polymeric Micelles
4.1.2. Liposomes/Niosomes
4.1.3. Nanocrystals
4.2. In Silico Modeling
4.2.1. Polymeric Micelles
4.2.2. Liposomes/Transfersomes
4.2.3. Nanocrystals
4.3. Machine Learning
4.3.1. Polymeric Micelles
4.3.2. Liposomes/Niosomes
4.3.3. Nanocrystals
4.4. Digital Twins
5. Regulatory Issues
6. Future Prospects
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ANN | Artificial neural network |
| AI | Artificial intelligence |
| API | Active pharmaceutical ingredient |
| ASP | Antisolvent precipitation |
| BWM | Ball wet milling |
| CESS® | Controlled expansion of supercritical solution |
| CMAs | Critical material attributes |
| CNS | Central nervous system |
| CPPs | Critical process parameters |
| CQAs | Critical quality attributes |
| 3D | Three-dimensional |
| Dexi | Dexibuprofen |
| DMT | Disease-modifying treatment |
| DRZ | Dorzolamide hydrochloride |
| EAE | Experimental autoimmune encephalomyelitis |
| EUD | Eudragit |
| FD | Freeze-dried |
| FDA | Food and Drug Administration |
| GA | Glatiramer acetate |
| GI | Gastrointestinal |
| hERG | Human ether-a-go-go-related gene |
| HME | Hot-melt extrusion |
| HPH | High-pressure homogenization |
| HPMC | Hydroxypropyl methylcellulose |
| IOP | Intraocular pressure |
| ISGS | In situ gelling system |
| MD | Molecular dynamics |
| MIMO | Multiple-input–multiple-output |
| MISO | Multiple-input–single-output |
| MF | Microfluidic technology |
| ML | Machine learning |
| MRE | Mean relative error |
| MS | Multiple sclerosis |
| NanoPRX | Nanoformed piroxicam |
| NN | Neural network |
| NAP | Naproxen |
| NBCD | Non-biological complex drug |
| PEO-PPO-PEO | Poly(ethylene oxide)-Poly(propylene oxide)-Poly(ethylene oxide) |
| PAL | Palmatine HCL |
| PAT | Process analytical technology |
| PDI | Polydispersity index |
| PVP | Polyvinyl pyrrolidone |
| QbD | Quality by design |
| RTD | Room temperature-dried |
| SDS | Sodium dodecyl sulfate |
| SVM | Support vector machines |
| SSE | Semi-solid extrusion |
| TEER | Transepithelial electrical resistance |
| TIM | Timolol maleate |
| TNF-α | Tumor necroses factor-α |
References
- Hariry, R.E.; Barenji, R.V.; Paradkar, A. Towards Pharma 4.0 in clinical trials: A future-orientated perspective. Drug Discov. Today 2022, 27, 315–325. [Google Scholar] [CrossRef]
- Sharma, M.; Sehrawat, R.; Luthra, S.; Daim, T.; Bakry, D. Moving Towards Industry 5.0 in the Pharmaceutical Manufacturing Sector: Challenges and Solutions for Germany. IEEE Trans. Eng. Manag. 2022, 1–18. [Google Scholar] [CrossRef]
- Arden, N.S.; Fisher, A.C.; Tyner, K.; Yu, L.X.; Lee, S.L.; Kopcha, M. Industry 4.0 for pharmaceutical manufacturing: Preparing for the smart factories of the future. Int. J. Pharm. 2021, 602, 120554. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Patel, P.; Shah, M. A comprehensive study on Industry 4.0 in the pharmaceutical industry for sustainable development. Environ. Sci. Pollut. Res. 2023, 30, 90088. [Google Scholar] [CrossRef] [PubMed]
- Grangeia, H.B.; Silva, C.; Simões, S.P.; Reis, M.S. Quality by design in pharmaceutical manufacturing: A systematic review of current status, challenges and future perspectives. Eur. J. Pharm. Biopharm. 2020, 147, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Inuwa, H.M.; Ravi Raja, A.; Kumar, A.; Singh, B.; Singh, S. Status of Industry 4.0 applications in healthcare 4.0 and Pharma 4.0. Mater. Today Proc. 2022, 62, 3593–3598. [Google Scholar] [CrossRef]
- Wang, W.; Ye, Z.; Gao, H.; Ouyang, D. Computational pharmaceutics—A new paradigm of drug delivery. J. Control. Release 2021, 338, 119–136. [Google Scholar] [CrossRef]
- Mak, K.K.; Pichika, M.R. Artificial intelligence in drug development: Present status and future prospects. Drug Discov. Today 2019, 24, 773–780. [Google Scholar] [CrossRef]
- Paul, D.; Sanap, G.; Shenoy, S.; Kalyane, D.; Kalia, K.; Tekade, R.K. Artificial intelligence in drug discovery and development. Drug Discov. Today 2021, 26, 80–93. [Google Scholar] [CrossRef]
- Rocco, P.; Musazzi, U.M.; Franzè, S.; Minghetti, P. Copies of nonbiological complex drugs: Generic, hybrid or biosimilar? Drug Discov. Today 2019, 24, 250–255. [Google Scholar] [CrossRef]
- Zagalo, D.M.; Sousa, J.; Simões, S. Quality by design (QbD) approach in marketing authorization procedures of Non-Biological Complex Drugs: A critical evaluation. Eur. J. Pharm. Biopharm. 2022, 178, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Saha, E.; Rathore, P.; Parida, R.; Rana, N.P. The interplay of emerging technologies in pharmaceutical supply chain performance: An empirical investigation for the rise of Pharma 4.0. Technol. Forecast. Soc. Chang. 2022, 181, 121768. [Google Scholar] [CrossRef]
- Ragelle, H.; Rahimian, S.; Guzzi, E.A.; Westenskow, P.D.; Tibbitt, M.W.; Schwach, G.; Langer, R. Additive manufacturing in drug delivery: Innovative drug product design and opportunities for industrial application. Adv. Drug Deliv. Rev. 2021, 178, 113990. [Google Scholar] [CrossRef] [PubMed]
- Muñiz Castro, B.; Elbadawi, M.; Ong, J.J.; Pollard, T.; Song, Z.; Gaisford, S.; Pérez, G.; Basit, A.W.; Cabalar, P.; Goyanes, A. Machine learning predicts 3D printing performance of over 900 drug delivery systems. J. Control. Release 2021, 337, 530–545. [Google Scholar] [CrossRef]
- Kotta, S.; Nair, A.; Alsabeelah, N. Davidopoulou & Ouranidis, 2022. Curr. Pharm. Des. 2018, 24, 5039–5048. [Google Scholar] [CrossRef]
- Jacob, S.; Nair, A.B.; Patel, V.; Shah, J. 3D Printing Technologies: Recent Development and Emerging Applications in Various Drug Delivery Systems. AAPS PharmSciTech 2020, 21, 220. [Google Scholar] [CrossRef]
- Tracy, T.; Wu, L.; Liu, X.; Cheng, S.; Li, X. 3D printing: Innovative solutions for patients and pharmaceutical industry. Int. J. Pharm. 2023, 631, 122480. [Google Scholar] [CrossRef]
- Awad, A.; Yao, A.; Trenfield, S.J.; Goyanes, A.; Gaisford, S.; Basit, A.W. 3D printed tablets (Printlets) with braille and moon patterns for visually impaired patients. Pharmaceutics 2020, 12, 172. [Google Scholar] [CrossRef]
- Awad, A.; Fina, F.; Trenfield, S.J.; Patel, P.; Goyanes, A.; Gaisford, S.; Basit, A.W. 3D printed pellets (Miniprintlets): A novel, multi-drug, controlled release platform technology. Pharmaceutics 2019, 11, 148. [Google Scholar] [CrossRef]
- Melocchi, A.; Uboldi, M.; Briatico-Vangosa, F.; Moutaharrik, S.; Cerea, M.; Foppoli, A.; Maroni, A.; Palugan, L.; Zema, L.; Gazzaniga, A. The chronotopicTM system for pulsatile and colonic delivery of active molecules in the era of precision medicine: Feasibility by 3D printing via fused deposition modeling (fdm). Pharmaceutics 2021, 13, 759. [Google Scholar] [CrossRef]
- Picco, C.J.; Utomo, E.; McClean, A.; Domínguez-Robles, J.; Anjani, Q.K.; Volpe-Zanutto, F.; McKenna, P.E.; Acheson, J.G.; Malinova, D.; Donnelly, R.F.; et al. Development of 3D-printed subcutaneous implants using concentrated polymer/drug solutions. Int. J. Pharm. 2023, 631, 122477. [Google Scholar] [CrossRef] [PubMed]
- Charoenying, T.; Patrojanasophon, P.; Ngawhirunpat, T.; Rojanarata, T.; Akkaramongkolporn, P.; Opanasopit, P. Fabrication of floating capsule-in- 3D-printed devices as gastro-retentive delivery systems of amoxicillin. J. Drug Deliv. Sci. Technol. 2020, 55, 101393. [Google Scholar] [CrossRef]
- Viceconti, M.; Emili, L.; Afshari, P.; Courcelles, E.; Curreli, C.; Famaey, N.; Geris, L.; Horner, M.; Jori, M.C.; Kulesza, A.; et al. Possible Contexts of Use for in Silico Trials Methodologies: A Consensus-Based Review. IEEE J. Biomed. Health Inform. 2021, 25, 3977–3982. [Google Scholar] [CrossRef] [PubMed]
- Brodland, G.W. How computational models can help unlock biological systems. Semin. Cell Dev. Biol. 2015, 47–48, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.; Bang, S.; Son, J.; Kim, D.; Jeong, Y.; Kim, P.; Yang, J.; Eom, J.H.; Choi, N.; Kim, H.N. Brain physiome: A concept bridging in vitro 3D brain models and in silico models for predicting drug toxicity in the brain. Bioact. Mater. 2022, 13, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, M.; Ellison, C.M.; Cronin, M.T.D.; Pastor, M.; Steger-Hartmann, T.; Munoz-Muriendas, J.; Pognan, F.; Madden, J.C. Ensuring confidence in predictions: A scheme to assess the scientific validity of in silico models. Adv. Drug Deliv. Rev. 2015, 86, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Ai, N.; Fan, X.; Ekins, S. In silico methods for predicting drug-drug interactions with cytochrome P-450s, transporters and beyond. Adv. Drug Deliv. Rev. 2015, 86, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Montanari, F.; Ecker, G.F. Prediction of drug-ABC-transporter interaction—Recent advances and future challenges. Adv. Drug Deliv. Rev. 2015, 86, 17–26. [Google Scholar] [CrossRef]
- Sung, B. In silico modeling of endocrine organ-on-a-chip systems. Math. Biosci. 2022, 352, 108900. [Google Scholar] [CrossRef]
- Tian, S.; Wang, J.; Li, Y.; Li, D.; Xu, L.; Hou, T. The application of in silico drug-likeness predictions in pharmaceutical research. Adv. Drug Deliv. Rev. 2015, 86, 2–10. [Google Scholar] [CrossRef]
- Villoutreix, B.O.; Taboureau, O. Computational investigations of hERG channel blockers: New insights and current predictive models. Adv. Drug Deliv. Rev. 2015, 86, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Bannigan, P.; Aldeghi, M.; Bao, Z.; Häse, F.; Aspuru-Guzik, A.; Allen, C. Machine learning directed drug formulation development. Adv. Drug Deliv. Rev. 2021, 175, 113806. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Srivastava, D.; Sahu, M.; Tiwari, S.; Ambasta, R.K.; Kumar, P. Artificial Intelligence to Deep Learning: Machine Intelligence Approach for Drug Discovery; Springer: Berlin/Heidelberg, Germany, 2021; Volume 25, ISBN 0123456789. [Google Scholar]
- Dinh, A.; Miertschin, S.; Young, A.; Mohanty, S.D. A data-driven approach to predicting diabetes and cardiovascular disease with machine learning. BMC Med. Inform. Decis. Mak. 2019, 19, 211. [Google Scholar] [CrossRef] [PubMed]
- Eckhardt, C.M.; Madjarova, S.J.; Williams, R.J.; Ollivier, M.; Karlsson, J.; Pareek, A.; Nwachukwu, B.U. Unsupervised machine learning methods and emerging applications in healthcare. Knee Surg. Sport. Traumatol. Arthrosc. 2023, 31, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Dedeloudi, A.; Weaver, E.; Lamprou, D.A. Machine learning in additive manufacturing & Microfluidics for smarter and safer drug delivery systems. Int. J. Pharm. 2023, 636, 122818. [Google Scholar] [CrossRef]
- Chou, K.C.; Cai, Y.D. Using functional domain composition and support vector machines for prediction of protein subcellular location. J. Biol. Chem. 2002, 277, 45765–45769. [Google Scholar] [CrossRef]
- Cabrera, A.; Bouterse, A.; Nelson, M.; Razzouk, J.; Ramos, O.; Chung, D.; Cheng, W.; Danisa, O. Use of random forest machine learning algorithm to predict short term outcomes following posterior cervical decompression with instrumented fusion. J. Clin. Neurosci. 2023, 107, 167–171. [Google Scholar] [CrossRef]
- Schütt, M.; Stamatopoulos, K.; Batchelor, H.K.; Simmons, M.J.H.; Alexiadis, A. Development of a digital twin of a tablet that mimics a real solid dosage form: Differences in the dissolution profile in conventional mini-USP II and a biorelevant colon model. Eur. J. Pharm. Sci. 2022, 179, 106310. [Google Scholar] [CrossRef]
- Moreno-Benito, M.; Lee, K.T.; Kaydanov, D.; Verrier, H.M.; Blackwood, D.O.; Doshi, P. Digital twin of a continuous direct compression line for drug product and process design using a hybrid flowsheet modelling approach. Int. J. Pharm. 2022, 628, 122336. [Google Scholar] [CrossRef]
- Davidopoulou, C.; Ouranidis, A. Pharma 4.0-Artificially Intelligent Digital Twins for Solidified Nanosuspensions. Pharmaceutics 2022, 14, 2113. [Google Scholar] [CrossRef]
- Schmidt, A.; Helgers, H.; Vetter, F.L.; Juckers, A.; Strube, J. Digital Twin of mRNA-Based SARS-COVID-19 Vaccine Real-Time Release Testing. Processes 2021, 9, 748. [Google Scholar] [CrossRef]
- Sun, Z.; Jiang, J.; Chen, X. Evaluation of Therapeutic Equivalence for the Follow-On Version of Intravenously Administered Non-Biological Complex Drugs. Clin. Pharmacokinet. 2020, 59, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Klein, K.; Stolk, P.; De Bruin, M.L.; Leufkens, H.G.M.; Crommelin, D.J.A.; De Vlieger, J.S.B. The EU regulatory landscape of non-biological complex drugs (NBCDs) follow-on products: Observations and recommendations. Eur. J. Pharm. Sci. 2019, 133, 228–235. [Google Scholar] [CrossRef]
- Crommelin, D.J.A.; Shah, V.P.; Klebovich, I.; Mcneil, S.E.; Weinstein, V.; Flühmann, B.; Mühlebach, S.; Vlieger, J.S.B. The similarity question for biologicals and non-biological complex drugs. Eur. J. Pharm. Sci. 2015, 76, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, R.S.; Silva-Lima, B.; Magro, F.; Alcobia, A.; da Costa, F.L.; Feio, J. Non-biological Complex Drugs (NBCDs): Complex Pharmaceuticals in Need of Individual Robust Clinical Assessment Before Any Therapeutic Equivalence Decision. Front. Med. 2020, 7, 590527. [Google Scholar] [CrossRef] [PubMed]
- Schellekens, H.; Klinger, E.; Mühlebach, S.; Brin, J.F.; Storm, G.; Crommelin, D.J.A. The therapeutic equivalence of complex drugs. Regul. Toxicol. Pharmacol. 2011, 59, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Ghezzi, M.; Pescina, S.; Padula, C.; Santi, P.; Del Favero, E.; Cantù, L.; Nicoli, S. Polymeric micelles in drug delivery: An insight of the techniques for their characterization and assessment in biorelevant conditions. J. Control. Release 2021, 332, 312–336. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.; Ramsey, J.D.; Kabanov, A.V. Polymeric micelles for the delivery of poorly soluble drugs: From nanoformulation to clinical approval. Adv. Drug Deliv. Rev. 2020, 156, 80–118. [Google Scholar] [CrossRef]
- He, Z.; Wan, X.; Schulz, A.; Bludau, H.; Dobrovolskaia, M.A.; Stern, S.T.; Montgomery, S.A.; Yuan, H.; Li, Z.; Alakhova, D.; et al. A high capacity polymeric micelle of paclitaxel: Implication of high dose drug therapy to safety and in vivo anti-cancer activity. Biomaterials 2016, 101, 296–309. [Google Scholar] [CrossRef]
- Ghosh, B.; Biswas, S. Polymeric micelles in cancer therapy: State of the art. J. Control. Release 2021, 332, 127–147. [Google Scholar] [CrossRef]
- Cagel, M.; Tesan, F.C.; Bernabeu, E.; Salgueiro, M.J.; Zubillaga, M.B.; Moretton, M.A.; Chiappetta, D.A. Polymeric mixed micelles as nanomedicines: Achievements and perspectives. Eur. J. Pharm. Biopharm. 2017, 113, 211–228. [Google Scholar] [CrossRef] [PubMed]
- Ebrahim Attia, A.B.; Ong, Z.Y.; Hedrick, J.L.; Lee, P.P.; Ee, P.L.R.; Hammond, P.T.; Yang, Y.Y. Mixed micelles self-assembled from block copolymers for drug delivery. Curr. Opin. Colloid Interface Sci. 2011, 16, 182–194. [Google Scholar] [CrossRef]
- Guimarães, D.; Cavaco-Paulo, A.; Nogueira, E. Design of liposomes as drug delivery system for therapeutic applications. Int. J. Pharm. 2021, 601, 120571. [Google Scholar] [CrossRef] [PubMed]
- Akbarzadeh, A.; Rezaei-sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N. Liposome: Classification, preparation, and applications. Nanoscale Res. Lett. 2013, 22, 102. [Google Scholar] [CrossRef] [PubMed]
- Jensen, G.M.; Hodgson, D.F. Opportunities and challenges in commercial pharmaceutical liposome applications. Adv. Drug Deliv. Rev. 2020, 154–155, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Saraf, S.; Jain, A.; Tiwari, A.; Verma, A.; Panda, P.K.; Jain, S.K. Advances in liposomal drug delivery to cancer: An overview. J. Drug Deliv. Sci. Technol. 2020, 56, 101549. [Google Scholar] [CrossRef]
- Atallah, C.; Viennet, C.; Robin, S.; Ibazizen, S.; Greige-Gerges, H.; Charcosset, C. Effect of cysteamine hydrochloride-loaded liposomes on skin depigmenting and penetration. Eur. J. Pharm. Sci. 2022, 168, 106082. [Google Scholar] [CrossRef]
- Costa, R.; Santos, L. Delivery systems for cosmetics—From manufacturing to the skin of natural antioxidants. Powder Technol. 2017, 322, 402–416. [Google Scholar] [CrossRef]
- Pando, D.; Matos, M.; Gutiérrez, G.; Pazos, C. Formulation of resveratrol entrapped niosomes for topical use. Colloids Surfaces B Biointerfaces 2015, 128, 398–404. [Google Scholar] [CrossRef]
- Chen, S.; Hanning, S.; Falconer, J.; Locke, M.; Wen, J. Recent advances in non-ionic surfactant vesicles (niosomes): Fabrication, characterization, pharmaceutical and cosmetic applications. Eur. J. Pharm. Biopharm. 2019, 144, 18–39. [Google Scholar] [CrossRef]
- Aparajay, P.; Dev, A. Functionalized niosomes as a smart delivery device in cancer and fungal infection. Eur. J. Pharm. Sci. 2022, 168, 106052. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, H.; Kohli, K.; Kumar, V.K. Nano-transfersomes as a novel carrier for transdermal delivery. Int. J. Pharm. 2013, 454, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Lei, W.; Yu, C.; Lin, H.; Zhou, X. Development of tacrolimus-loaded transfersomes for deeper skin penetration enhancement and therapeutic effect improvement in vivo. Asian J. Pharm. Sci. 2013, 8, 336–345. [Google Scholar] [CrossRef]
- Al Shuwaili, A.H.; Rasool, B.K.A.; Abdulrasool, A.A. Optimization of elastic transfersomes formulations for transdermal delivery of pentoxifylline. Eur. J. Pharm. Biopharm. 2016, 102, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Rocco, P.; Eberini, I.; Musazzi, U.M.; Franzè, S.; Minghetti, P. Glatiramer acetate: A complex drug beyond biologics. Eur. J. Pharm. Sci. 2019, 133, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Weinstock-Guttman, B.; Nair, K.V.; Glajch, J.L.; Ganguly, T.C.; Kantor, D. Two decades of glatiramer acetate: From initial discovery to the current development of generics. J. Neurol. Sci. 2017, 376, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Melamed-Gal, S.; Loupe, P.; Timan, B.; Weinstein, V.; Kolitz, S.; Zhang, J.; Funt, J.; Komlosh, A.; Ashkenazi, N.; Bar-Ilan, O.; et al. Physicochemical, biological, functional and toxicological characterization of the European follow-on glatiramer acetate product as compared with Copaxone. eNeurologicalSci 2018, 12, 19–30. [Google Scholar] [CrossRef]
- Nikravesh, N.; Borchard, G.; Hofmann, H.; Philipp, E.; Flühmann, B.; Wick, P. Factors influencing safety and efficacy of intravenous iron-carbohydrate nanomedicines: From production to clinical practice. Nanomed. Nanotechnol. Biol. Med. 2020, 26, 102178. [Google Scholar] [CrossRef]
- Percy, L.; Mansour, D.; Fraser, I.; Hon, F. Iron deficiency and iron deficiency anaemia in women. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 40, 55–67. [Google Scholar] [CrossRef]
- Auerbach, M.; Ballard, H. Clinical use of intravenous iron: Administration, efficacy, and safety. Hematol. Am. Soc. Hematol. Educ. Program 2010, 2010, 338–347. [Google Scholar] [CrossRef]
- Zheng, N.; Sun, D.D.; Zou, P.; Jiang, W. Scientific and Regulatory Considerations for Generic Complex Drug Products Containing Nanomaterials. AAPS J. 2017, 19, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Müller, R.H.; Gohla, S.; Keck, C.M. State of the art of nanocrystals—Special features, production, nanotoxicology aspects and intracellular delivery. Eur. J. Pharm. Biopharm. 2011, 78, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, I.S.; Hu, H.; Yin, L.; He, W. Drug nanocrystals: Fabrication methods and promising therapeutic applications. Int. J. Pharm. 2019, 562, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Yang, W.; Feng, T.; Lin, J.; Huang, P. Drug nanocrystals for cancer therapy. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2018, 10, e1499. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Gao, W.; Hu, H.; Ma, K.; He, B.; Dai, W.; Wang, X.; Wang, J.; Zhang, X.; Zhang, Q. Novel thermo-sensitive hydrogel system with paclitaxel nanocrystals: High drug-loading, sustained drug release and extended local retention guaranteeing better efficacy and lower toxicity. J. Control. Release 2014, 174, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Almeida, A.; Linares, V.; Mora-Castaño, G.; Casas, M.; Caraballo, I.; Sarmento, B. 3D printed systems for colon-specific delivery of camptothecin-loaded chitosan micelles. Eur. J. Pharm. Biopharm. 2021, 167, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Linares, V.; Casas, M.; Caraballo, I. Printfills: 3D printed systems combining fused deposition modeling and injection volume filling. Application to colon-specific drug delivery. Eur. J. Pharm. Biopharm. 2019, 134, 138–143. [Google Scholar] [CrossRef]
- Cisneros, K.; Chowdhury, N.; Coleman, E.; Ferdous, T.; Su, H.; Jennings, J.A.; Bumgardner, J.D.; Fujiwara, T. Long-Term Controlled Release of Simvastatin from Photoprinted Triple-Networked Hydrogels Composed of Modified Chitosan and PLA–PEG Micelles. Macromol. Biosci. 2021, 21, 2100123. [Google Scholar] [CrossRef]
- Sommonte, F.; Weaver, E.; Mathew, E.; Denora, N.; Lamprou, D.A. In-House Innovative “Diamond Shaped” 3D Printed Microfluidic Devices for Lysozyme-Loaded Liposomes. Pharmaceutics 2022, 14, 2484. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, L.; Xiang, S.; Jiang, C.; Wu, W.; Ruan, S.; Du, Q.; Chen, T.; Xue, Y.; Chen, H.; et al. Formulation and Characterization of a 3D-Printed Cryptotanshinone-Loaded Niosomal Hydrogel for Topical Therapy of Acne. AAPS PharmSciTech 2020, 21, 159. [Google Scholar] [CrossRef]
- Groetsch, A.; Stelzl, S.; Nagel, Y.; Kochetkova, T.; Scherrer, N.C.; Ovsianikov, A.; Michler, J.; Pethö, L.; Siqueira, G.; Nyström, G.; et al. Microscale 3D Printing and Tuning of Cellulose Nanocrystals Reinforced Polymer Nanocomposites. Small 2023, 19, e2202470. [Google Scholar] [CrossRef] [PubMed]
- Germini, G.; Peltonen, L. 3D Printing of Drug Nanocrystals for Film Formulations. Molecules 2021, 26, 3941. [Google Scholar] [CrossRef] [PubMed]
- Phan, V.H.G.; Murugesan, M.; Huong, H.; Le, T.T.; Phan, T.H.; Manivasagan, P.; Mathiyalagan, R.; Jang, E.S.; Yang, D.C.; Li, Y.; et al. Cellulose Nanocrystals-Incorporated Thermosensitive Hydrogel for Controlled Release, 3D Printing, and Breast Cancer Treatment Applications. ACS Appl. Mater. Interfaces 2022, 14, 42812–42826. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Vidal, L.; Real, J.P.; Real, D.A.; Camacho, N.; Kogan, M.J.; Paredes, A.J.; Palma, S.D. Nanocrystal-based 3D-printed tablets: Semi-solid extrusion using melting solidification printing process (MESO-PP) for oral administration of poorly soluble drugs. Int. J. Pharm. 2022, 611, 121311. [Google Scholar] [CrossRef] [PubMed]
- Tambe, S.M.; Jain, D.D.; Hasmukh Mehta, C.; Ashwini, T.; Yogendra Nayak, U.; Amin, P.D. Hot-melt extruded in situ gelling systems (MeltDrops Technology): Formulation development, in silico modelling and in vivo studies. Eur. J. Pharm. Biopharm. 2023, 188, 108–124. [Google Scholar] [CrossRef] [PubMed]
- Duran, T.; Costa, A.; Gupta, A.; Xu, X.; Zhang, H.; Burgess, D.; Chaudhuri, B. Coarse-Grained Molecular Dynamics Simulations of Paclitaxel-Loaded Polymeric Micelles. Mol. Pharm. 2022, 19, 1117–1134. [Google Scholar] [CrossRef]
- Bhendale, M.; Singh, J.K. Molecular Insights on Morphology, Composition, and Stability of Mixed Micelles Formed by Ionic Surfactant and Nonionic Block Copolymer in Water Using Coarse-Grained Molecular Dynamics Simulations. Langmuir 2023, 39, 5031–5040. [Google Scholar] [CrossRef]
- Ramajayam, K.K.; Wolfe, A.M.; Motamarry, A.; Yost, J.; Yost, M.J.; Haemmerich, D. Computer Modeling of Drug Delivery with Thermosensitive Liposomes in a Realistic Three-Dimensional Geometry. Proc. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. EMBS 2020, 2020, 5021–5024. [Google Scholar] [CrossRef]
- Cern, A.; Marcus, D.; Tropsha, A.; Barenholz, Y.; Goldblum, A. New drug candidates for liposomal delivery identified by computer modeling of liposomes’ remote loading and leakage. J. Control. Release 2017, 252, 18–27. [Google Scholar] [CrossRef]
- Bunker, A.; Magarkar, A.; Viitala, T. Rational design of liposomal drug delivery systems, a review: Combined experimental and computational studies of lipid membranes, liposomes and their PEGylation. Biochim. Biophys. Acta—Biomembr. 2016, 1858, 2334–2352. [Google Scholar] [CrossRef]
- Pathak, P.; Dhawan, V.; Magarkar, A.; Danne, R.; Govindarajan, S.; Ghosh, S.; Steiniger, F.; Chaudhari, P.; Gopal, V.; Bunker, A.; et al. Design of cholesterol arabinogalactan anchored liposomes for asialoglycoprotein receptor mediated targeting to hepatocellular carcinoma: In silico modeling, in vitro and in vivo evaluation. Int. J. Pharm. 2016, 509, 149–158. [Google Scholar] [CrossRef]
- Wu, X.; Dai, X.; Liao, Y.; Sheng, M.; Shi, X. Investigation on drug entrapment location in liposomes and transfersomes based on molecular dynamics simulation. J. Mol. Model. 2021, 27, 111. [Google Scholar] [CrossRef] [PubMed]
- Jablonka, L.; Ashtikar, M.; Gao, G.; Jung, F.; Thurn, M.; Preuß, A.; Scheglmann, D.; Albrecht, V.; Röder, B.; Wacker, M.G. Advanced in silico modeling explains pharmacokinetics and biodistribution of temoporfin nanocrystals in humans. J. Control. Release 2019, 308, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Ullah, N.; Khan, S.; Ahmed, S.; Govender, T.; Faidah, H.S.; De Matas, M.; Shahid, M.; Minhas, M.U.; Sohail, M.; Khurram, M. Dexibuprofen nanocrystals with improved therapeutic performance: Fabrication, characterization, in silico modeling, and in vivo evaluation. Int. J. Nanomed. 2018, 13, 1677–1692. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.E.; Ghandehari, H.; Facelli, J.C. A review of the applications of data mining and machine learning for the prediction of biomedical properties of nanoparticles. Comput. Methods Programs Biomed. 2016, 132, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Husseini, G.A.; Mjalli, F.S.; Pitt, W.G.; Abdel-Jabbar, N.M. Using ANN and MPC to optimize acoustically assisted Doxorubicin release from micelles. Proc. IASTED Int. Conf. Model. Simul. 2009, 8, 320–325. [Google Scholar]
- Han, R.; Ye, Z.; Zhang, Y.; Cheng, Y.; Zheng, Y.; Ouyang, D. Predicting liposome formulations by the integrated machine learning and molecular modeling approaches. Asian J. Pharm. Sci. 2023, 18, 100811. [Google Scholar] [CrossRef] [PubMed]
- Kashani-Asadi-Jafari, F.; Aftab, A.; Ghaemmaghami, S. A machine learning framework for predicting entrapment efficiency in niosomal particles. Int. J. Pharm. 2022, 627, 122203. [Google Scholar] [CrossRef]
- Subramanian, N.; Yajnik, A.; Murthy, R.S.R. Artificial neural network as an alternative to multiple regression analysis in optimizing formulation parameters of cytarabine liposomes. AAPS PharmSciTech 2004, 5, E4. [Google Scholar] [CrossRef]
- Rebollo, R.; Oyoun, F.; Corvis, Y.; El-Hammadi, M.M.; Saubamea, B.; Andrieux, K.; Mignet, N.; Alhareth, K. Microfluidic Manufacturing of Liposomes: Development and Optimization by Design of Experiment and Machine Learning. ACS Appl. Mater. Interfaces 2022, 14, 39736–39745. [Google Scholar] [CrossRef]
- Baptista, D.; Correia, J.; Pereira, B.; Rocha, M. Evaluating molecular representations in machine learning models for drug response prediction and interpretability. J. Integr. Bioinform. 2022, 19, 20220006. [Google Scholar] [CrossRef] [PubMed]
- Sansare, S.; Duran, T.; Mohammadiarani, H.; Goyal, M.; Yenduri, G.; Costa, A.; Xu, X.; O’Connor, T.; Burgess, D.; Chaudhuri, B. Artificial neural networks in tandem with molecular descriptors as predictive tools for continuous liposome manufacturing. Int. J. Pharm. 2021, 603, 120713. [Google Scholar] [CrossRef]
- Rathore, A.S.; Nikita, S.; Thakur, G.; Mishra, S. Artificial intelligence and machine learning applications in biopharmaceutical manufacturing. Trends Biotechnol. 2023, 41, 497–510. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ye, Z.; Liu, X.; Wei, Z.; Qiu, F.; Li, H.F.; Zheng, Y.; Ouyang, D. Can machine learning predict drug nanocrystals? J. Control. Release 2020, 322, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Attaran, M.; Attaran, S.; Celik, B.G. The impact of digital twins on the evolution of intelligent manufacturing and Industry 4.0. Adv. Comput. Intell. 2023, 3. [Google Scholar] [CrossRef] [PubMed]
- Trenfield, S.J.; Awad, A.; McCoubrey, L.E.; Elbadawi, M.; Goyanes, A.; Gaisford, S.; Basit, A.W. Advancing pharmacy and healthcare with virtual digital technologies. Adv. Drug Deliv. Rev. 2022, 182, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Pravin, S.; Sudhir, A. Integration of 3D printing with dosage forms: A new perspective for modern healthcare. Biomed. Pharmacother. 2018, 107, 146–154. [Google Scholar] [CrossRef]
- Bom, S.; Martins, A.M.; Ribeiro, H.M.; Marto, J. Diving into 3D (bio)printing: A revolutionary tool to customize the production of drug and cell-based systems for skin delivery. Int. J. Pharm. 2021, 605, 120794. [Google Scholar] [CrossRef]
- Jamróz, W.; Szafraniec, J.; Kurek, M.; Jachowicz, R. 3D Printing in Pharmaceutical and Medical Applications—Recent Achievements and Challenges. Pharm. Res. 2018, 35, 176. [Google Scholar] [CrossRef]
- Preis, M.; Öblom, H. 3D-Printed Drugs for Children—Are We Ready Yet? AAPS PharmSciTech 2017, 18, 303–308. [Google Scholar] [CrossRef]
- Methods, N.S. Performance Metrics for the Comparative Analysis of Clinical Risk Prediction Models. Circ. Cardiovasc. Qual. Outcomes 2021, 14, 1076–1086. [Google Scholar] [CrossRef]
- Rácz, A.; Bajusz, D.; Héberger, K. Multi-Level Comparison of Machine Learning Classifiers and Their Performance Metrics. Molecules 2019, 24, 2811. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malheiro, V.; Duarte, J.; Veiga, F.; Mascarenhas-Melo, F. Exploiting Pharma 4.0 Technologies in the Non-Biological Complex Drugs Manufacturing: Innovations and Implications. Pharmaceutics 2023, 15, 2545. https://doi.org/10.3390/pharmaceutics15112545
Malheiro V, Duarte J, Veiga F, Mascarenhas-Melo F. Exploiting Pharma 4.0 Technologies in the Non-Biological Complex Drugs Manufacturing: Innovations and Implications. Pharmaceutics. 2023; 15(11):2545. https://doi.org/10.3390/pharmaceutics15112545
Chicago/Turabian StyleMalheiro, Vera, Joana Duarte, Francisco Veiga, and Filipa Mascarenhas-Melo. 2023. "Exploiting Pharma 4.0 Technologies in the Non-Biological Complex Drugs Manufacturing: Innovations and Implications" Pharmaceutics 15, no. 11: 2545. https://doi.org/10.3390/pharmaceutics15112545