Modulating the Nature of Ionizable Lipids and Number of Layers in Hyaluronan-Decorated Lipid Nanoparticles for In Vitro Delivery of RNAi
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
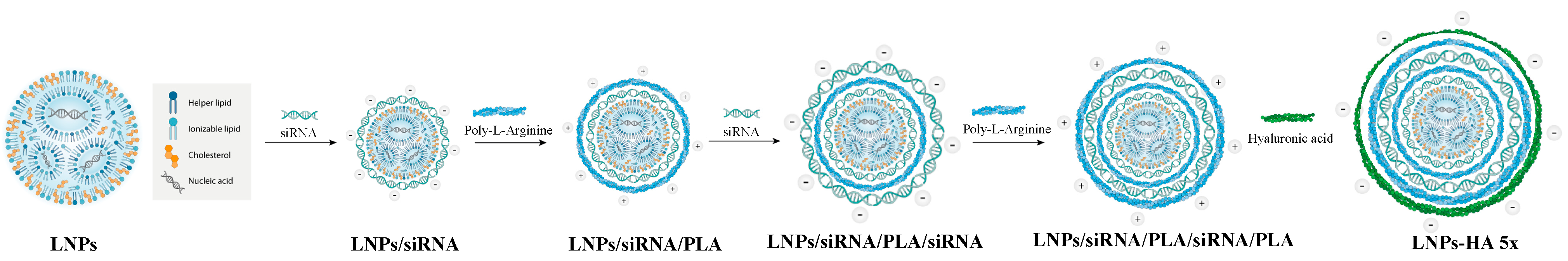
2.2. Synthesis and Characterization of HA-LNPs
2.3. Encapsulation Efficiency
2.4. Confocal Microscopy
2.5. Gene Transfection
2.6. Statistical Analysis
3. Results
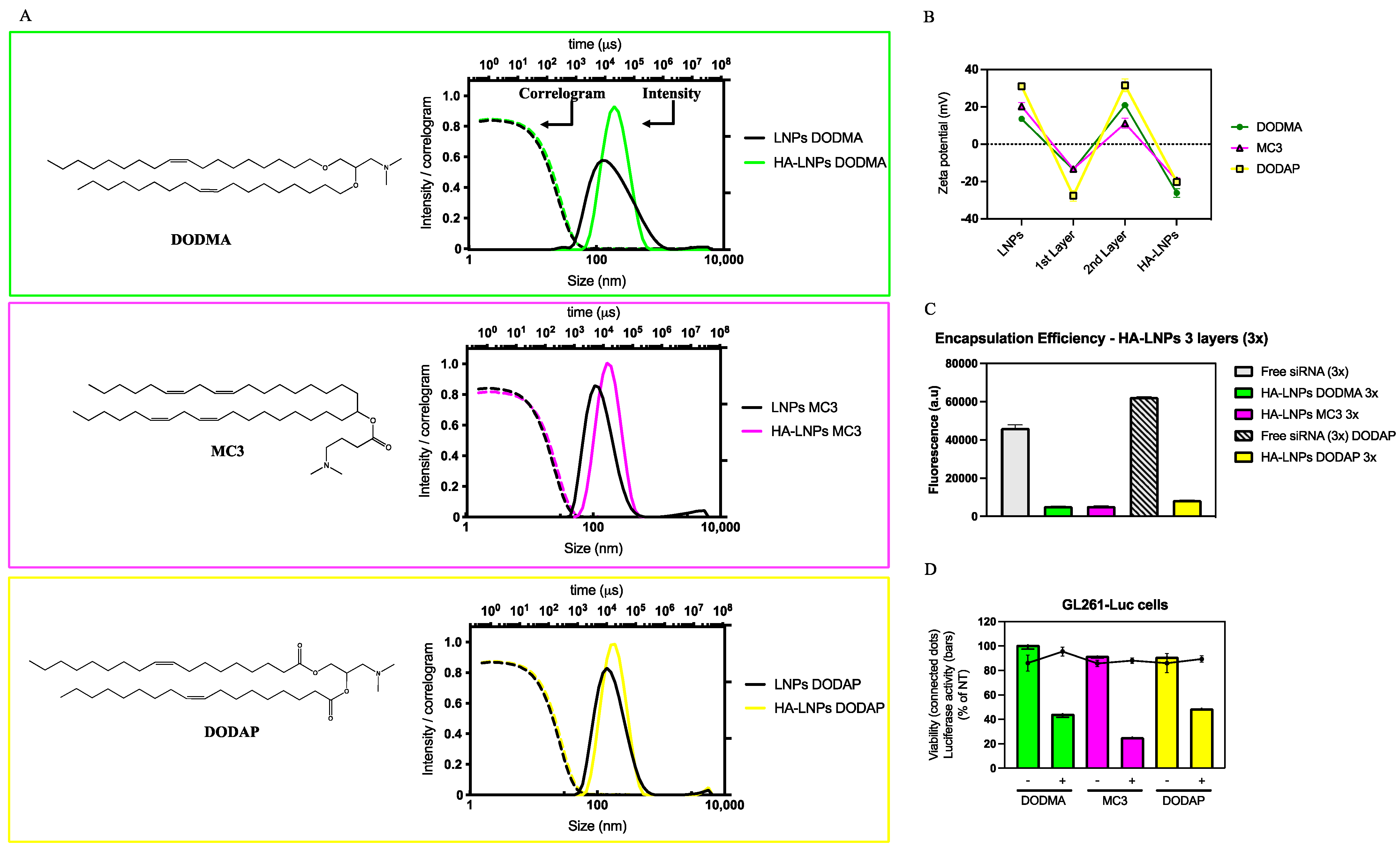
3.1. Formulation of HA-LNPs with Different Ionizable Lipids
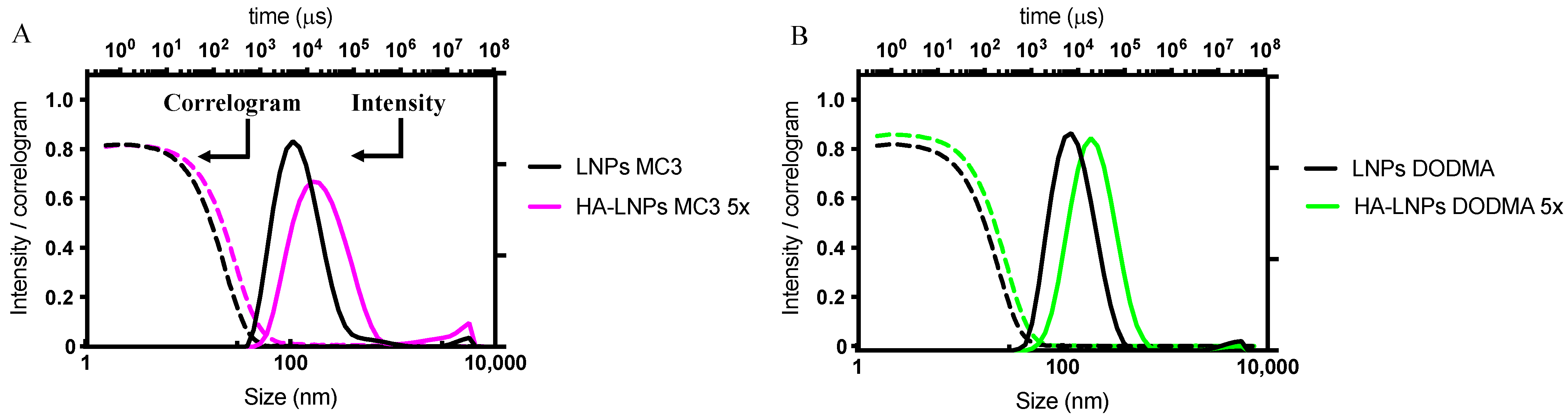
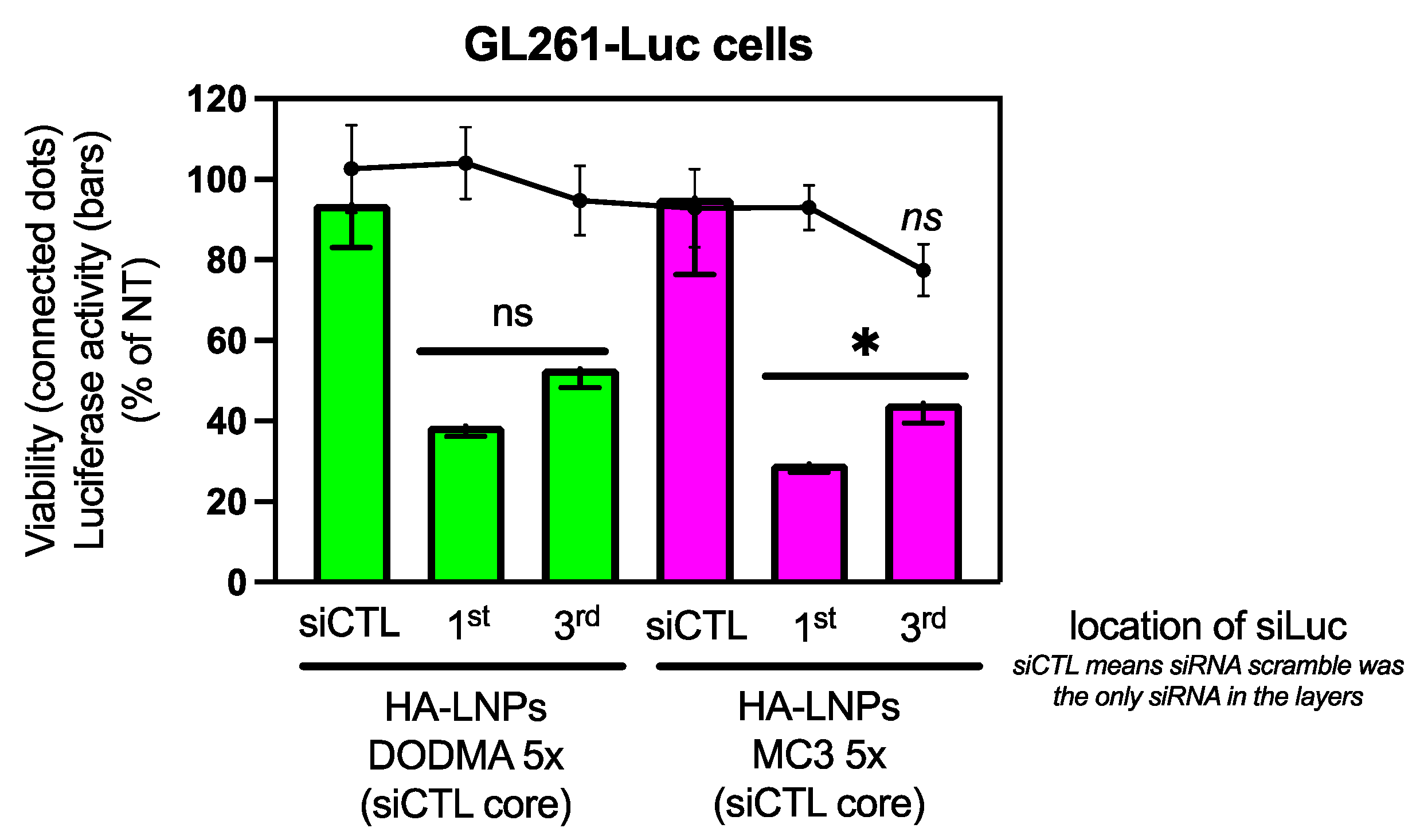
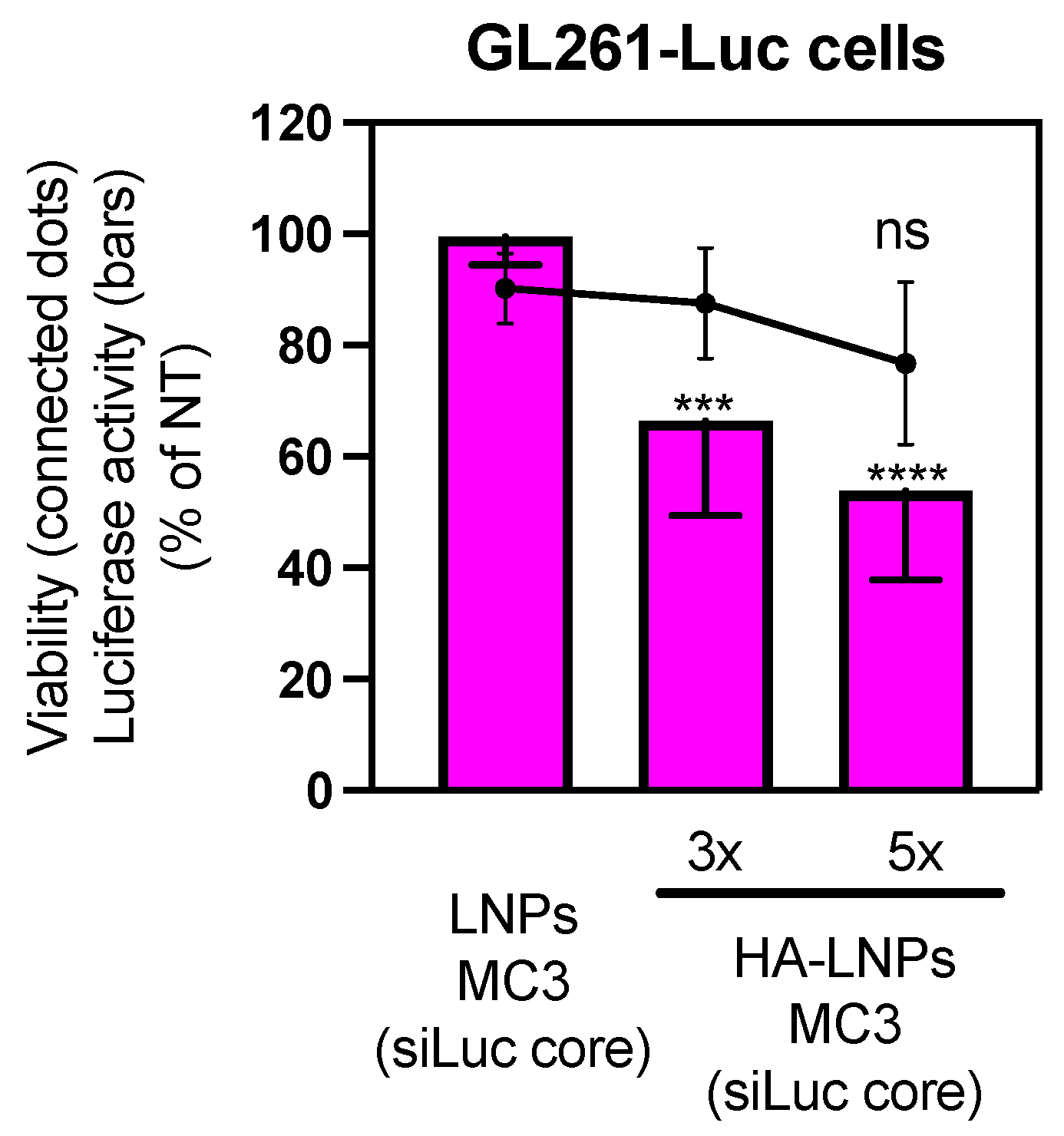
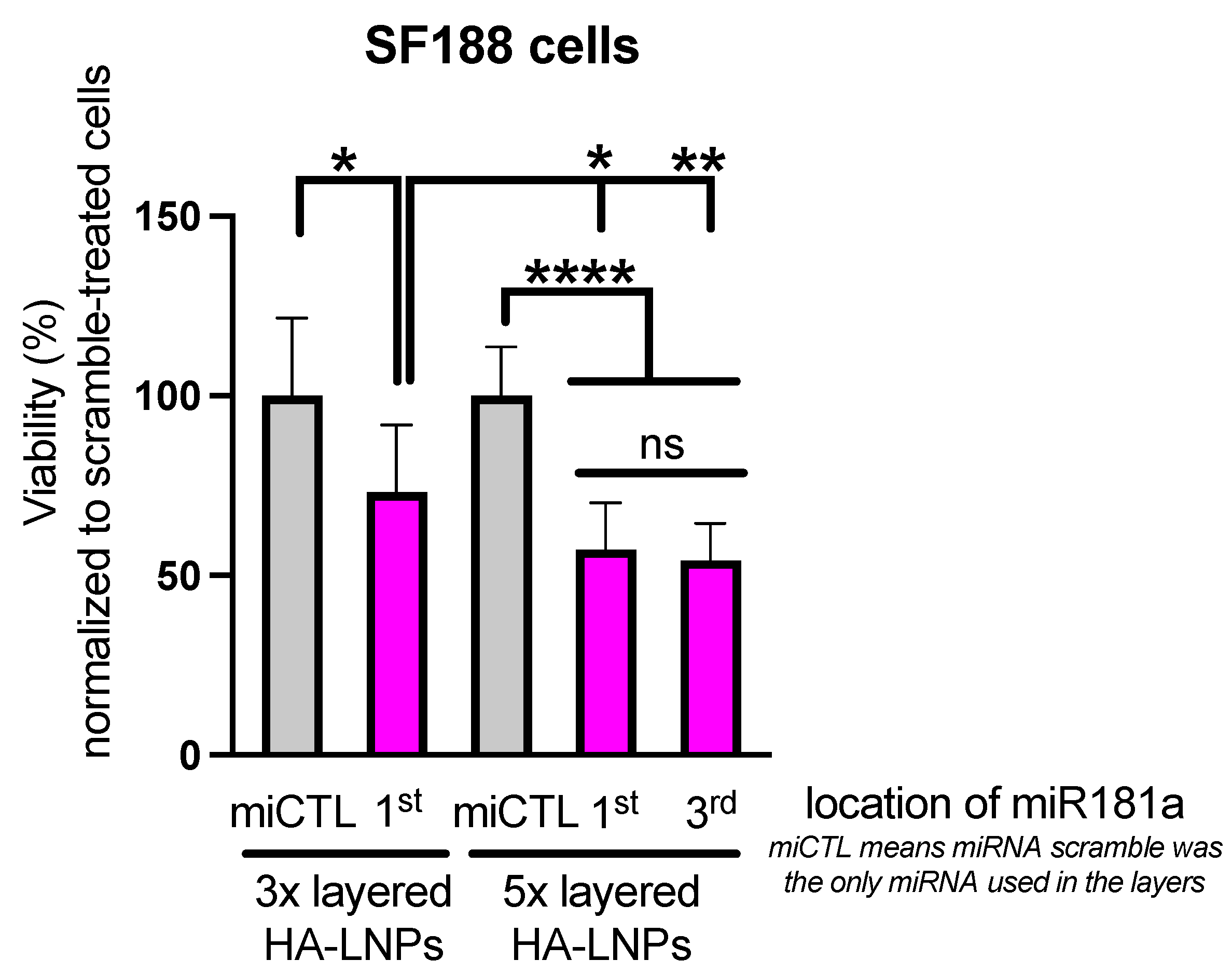
3.2. Construction of Five-Layered Hyaluronan-Decorated LNPs (HA-LNPs 5×)
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Younis, M.A.; Tawfeek, H.M.; Abdellatif, A.A.H.; Abdel-Aleem, J.A.; Harashima, H. Clinical translation of nanomedicines: Challenges, opportunities, and keys. Adv. Drug Deliv. Rev. 2022, 181, 114083. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Perez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Akinc, A.; Maier, M.A.; Manoharan, M.; Fitzgerald, K.; Jayaraman, M.; Barros, S.; Ansell, S.; Du, X.; Hope, M.J.; Madden, T.D.; et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019, 14, 1084–1087. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, K.A.; Langer, R.; Anderson, D.G. Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov. 2009, 8, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Ouyang, K.; Xu, X.; Xu, L.; Wen, C.; Zhou, X.; Qin, Z.; Xu, Z.; Sun, W.; Liang, Y. Nanoparticle Delivery of CRISPR/Cas9 for Genome Editing. Front. Genet. 2021, 12, 673286. [Google Scholar] [CrossRef] [PubMed]
- Pfeifle, A.; Thulasi Raman, S.N.; Lansdell, C.; Zhang, W.; Tamming, L.; Cecillon, J.; Laryea, E.; Patel, D.; Wu, J.; Gravel, C.; et al. DNA lipid nanoparticle vaccine targeting outer surface protein C affords protection against homologous Borrelia burgdorferi needle challenge in mice. Front. Immunol. 2023, 14, 1020134. [Google Scholar] [CrossRef] [PubMed]
- Tabatabaei, S.N.; Derbali, R.M.; Yang, C.; Superstein, R.; Hamel, P.; Chain, J.L.; Hardy, P. Co-delivery of miR-181a and melphalan by lipid nanoparticles for treatment of seeded retinoblastoma. J. Control Release 2019, 298, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Li, Y.; Bloomer, H.; Xu, Q. Developing Biodegradable Lipid Nanoparticles for Intracellular mRNA Delivery and Genome Editing. Acc. Chem. Res. 2021, 54, 4001–4011. [Google Scholar] [CrossRef]
- Love, K.T.; Mahon, K.P.; Levins, C.G.; Whitehead, K.A.; Querbes, W.; Dorkin, J.R.; Qin, J.; Cantley, W.; Qin, L.L.; Racie, T.; et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl. Acad. Sci. USA 2010, 107, 1864–1869. [Google Scholar] [CrossRef]
- Phan, H.T.; Passos Gibson, V.; Guedin, A.; Ibarboure, E.; El Mammeri, N.; Grelard, A.; Le Meins, J.F.; Dufourc, E.J.; Loquet, A.; Giasson, S.; et al. Switchable Lipids: From Conformational Switch to Macroscopic Changes in Lipid Vesicles. Langmuir 2023, 39, 3072–3082. [Google Scholar] [CrossRef] [PubMed]
- Passos Gibson, V.; Derbali, R.M.; Phan, H.T.; Tahiri, H.; Allen, C.; Hardy, P.; Chain, J.L. Survivin silencing improved the cytotoxicity of carboplatin and melphalan in Y79 and primary retinoblastoma cells. Int. J. Pharm. 2020, 589, 119824. [Google Scholar] [CrossRef] [PubMed]
- Viricel, W.; Poirier, S.; Mbarek, A.; Derbali, R.M.; Mayer, G.; Leblond, J. Cationic switchable lipids: pH-triggered molecular switch for siRNA delivery. Nanoscale 2017, 9, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Wei, T.; Farbiak, L.; Johnson, L.T.; Dilliard, S.A.; Siegwart, D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020, 15, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Hald Albertsen, C.; Kulkarni, J.A.; Witzigmann, D.; Lind, M.; Petersson, K.; Simonsen, J.B. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv. Drug Deliv. Rev. 2022, 188, 114416. [Google Scholar] [CrossRef] [PubMed]
- Melamed, J.R.; Yerneni, S.S.; Arral, M.L.; LoPresti, S.T.; Chaudhary, N.; Sehrawat, A.; Muramatsu, H.; Alameh, M.G.; Pardi, N.; Weissman, D.; et al. Ionizable lipid nanoparticles deliver mRNA to pancreatic beta cells via macrophage-mediated gene transfer. Sci. Adv. 2023, 9, eade1444. [Google Scholar] [CrossRef] [PubMed]
- Veiga, N.; Goldsmith, M.; Granot, Y.; Rosenblum, D.; Dammes, N.; Kedmi, R.; Ramishetti, S.; Peer, D. Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat. Commun. 2018, 9, 4493. [Google Scholar] [CrossRef] [PubMed]
- Alkekhia, D.; Hammond, P.T.; Shukla, A. Layer-by-Layer Biomaterials for Drug Delivery. Annu. Rev. Biomed. Eng. 2020, 22, 1–24. [Google Scholar] [CrossRef]
- Bishop, C.J.; Liu, A.L.; Lee, D.S.; Murdock, R.J.; Green, J.J. Layer-by-layer inorganic/polymeric nanoparticles for kinetically controlled multigene delivery. J. Biomed. Mater. Res. A 2016, 104, 707–713. [Google Scholar] [CrossRef]
- Boehnke, N.; Correa, S.; Hao, L.; Wang, W.; Straehla, J.P.; Bhatia, S.N.; Hammond, P.T. Theranostic Layer-by-Layer Nanoparticles for Simultaneous Tumor Detection and Gene Silencing. Angew. Chem. Int. Ed. Engl. 2020, 59, 2776–2783. [Google Scholar] [CrossRef]
- Choi, K.Y.; Correa, S.; Min, J.; Li, J.; Roy, S.; Laccetti, K.H.; Dreaden, E.; Kong, S.; Heo, R.; Roh, Y.H.; et al. Binary Targeting of siRNA to Hematologic Cancer Cells In Vivo using Layer-by-Layer Nanoparticles. Adv. Funct. Mater. 2019, 29, 1900018. [Google Scholar] [CrossRef] [PubMed]
- Correa, S.; Dreaden, E.C.; Gu, L.; Hammond, P.T. Engineering nanolayered particles for modular drug delivery. J. Control Release 2016, 240, 364–386. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.J.; Morton, S.W.; Ben-Akiva, E.; Dreaden, E.C.; Shopsowitz, K.E.; Hammond, P.T. Layer-by-layer nanoparticles for systemic codelivery of an anticancer drug and siRNA for potential triple-negative breast cancer treatment. ACS Nano 2013, 7, 9571–9584. [Google Scholar] [CrossRef]
- Gu, L.; Deng, Z.J.; Roy, S.; Hammond, P.T. A Combination RNAi-Chemotherapy Layer-by-Layer Nanoparticle for Systemic Targeting of KRAS/P53 with Cisplatin to Treat Non-Small Cell Lung Cancer. Clin. Cancer Res. 2017, 23, 7312–7323. [Google Scholar] [CrossRef] [PubMed]
- Guter, M.; Breunig, M. Layer-by-Layer Assembled Nanoparticles for siRNA Delivery. Methods Mol. Biol. 2019, 1943, 153–160. [Google Scholar]
- Kong, S.M.; Costa, D.F.; Jagielska, A.; Van Vliet, K.J.; Hammond, P.T. Stiffness of targeted layer-by-layer nanoparticles impacts elimination half-life, tumor accumulation, and tumor penetration. Proc. Natl. Acad. Sci. USA 2021, 118, e2104826118. [Google Scholar] [CrossRef]
- Yang, C.; Passos Gibson, V.; Hardy, P. The Role of MiR-181 Family Members in Endothelial Cell Dysfunction and Tumor Angiogenesis. Cells 2022, 11, 1670. [Google Scholar] [CrossRef]
- Passos Gibson, V.; Tahiri, H.; Yang, C.; Phan, Q.T.; Banquy, X.; Hardy, P. Hyaluronan decorated layer-by-layer assembled lipid nanoparticles for miR-181a delivery in glioblastoma treatment. Biomaterials 2023, 302, 122341. [Google Scholar] [CrossRef]
- Culty, M.; Nguyen, H.A.; Underhill, C.B. The hyaluronan receptor (CD44) participates in the uptake and degradation of hyaluronan. J. Cell Biol. 1992, 116, 1055–1062. [Google Scholar] [CrossRef]
- Cannava, C.; De Gaetano, F.; Stancanelli, R.; Venuti, V.; Paladini, G.; Caridi, F.; Ghica, C.; Crupi, V.; Majolino, D.; Ferlazzo, G.; et al. Chitosan-Hyaluronan Nanoparticles for Vinblastine Sulfate Delivery: Characterization and Internalization Studies on K-562 Cells. Pharmaceutics 2022, 14, 942. [Google Scholar] [CrossRef]
- Gaspar, R.; Coelho, F.; Silva, B.F.B. Lipid-Nucleic Acid Complexes: Physicochemical Aspects and Prospects for Cancer Treatment. Molecules 2020, 25, 5006. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Chen, S.; Cullis, P.R.; van der Meel, R. Lipid Nanoparticle Technology for Clinical Translation of siRNA Therapeutics. Acc. Chem. Res. 2019, 52, 2435–2444. [Google Scholar] [CrossRef] [PubMed]
- Nayef, L.; Castiello, R.; Tabrizian, M. Washless Method Enables Multilayer Coating of an Aggregation-Prone Nanoparticulate Drug Delivery System with Enhanced Yields, Colloidal Stability, and Scalability. Macromol. Biosci. 2017, 17, 1600535. [Google Scholar] [CrossRef]
- Guzman, E.; Mateos-Maroto, A.; Ruano, M.; Ortega, F.; Rubio, R.G. Layer-by-Layer polyelectrolyte assemblies for encapsulation and release of active compounds. Adv. Colloid. Interface Sci. 2017, 249, 290–307. [Google Scholar] [CrossRef]
- Poon, Z.; Lee, J.B.; Morton, S.W.; Hammond, P.T. Controlling in vivo stability and biodistribution in electrostatically assembled nanoparticles for systemic delivery. Nano Lett. 2011, 11, 2096–2103. [Google Scholar] [CrossRef]
- Lee, A.; Gosnell, N.; Milinkovic, D.; Taladriz-Blanco, P.; Rothen-Rutishauser, B.; Petri-Fink, A. Layer-by-Layer siRNA Particle Assemblies for Localized Delivery of siRNA to Epithelial Cells through Surface-Mediated Particle Uptake. ACS Appl. Bio Mater. 2023, 6, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, P.; Nuzzo, S.; Torino, E.; Condorelli, G.; Salvatore, M.; Grimaldi, A.M. Antifouling Strategies of Nanoparticles for Diagnostic and Therapeutic Application: A Systematic Review of the Literature. Nanomaterials 2021, 11, 780. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.M.; Cheng, T.L.; Roffler, S.R. Polyethylene Glycol Immunogenicity: Theoretical, Clinical, and Practical Aspects of Anti-Polyethylene Glycol Antibodies. ACS Nano 2021, 15, 14022–14048. [Google Scholar] [CrossRef]
- Dilliard, S.A.; Siegwart, D.J. Passive, active and endogenous organ-targeted lipid and polymer nanoparticles for delivery of genetic drugs. Nat. Rev. Mater. 2023, 8, 282–300. [Google Scholar] [CrossRef]
- Hoang Thi, T.T.; Pilkington, E.H.; Nguyen, D.H.; Lee, J.S.; Park, K.D.; Truong, N.P. The Importance of Poly(ethylene glycol) Alternatives for Overcoming PEG Immunogenicity in Drug Delivery and Bioconjugation. Polymers 2020, 12, 298. [Google Scholar] [CrossRef]
- Lamson, N.G.; Pickering, A.J.; Wyckoff, J.; Ganesh, P.; Calle, E.A.; Straehla, J.P.; Hammond, P.T. Trafficking through the blood-brain barrier is directed by core and outer surface components of layer-by-layer nanoparticles. Bioeng. Transl. Med. 2023. [Google Scholar] [CrossRef]
- Pickering, A.J.; Lamson, N.G.; Marand, M.H.; Hwang, W.; Straehla, J.P.; Hammond, P.T. Layer-by-Layer Polymer Functionalization Improves Nanoparticle Penetration and Glioblastoma Targeting in the Brain. ACS Nano 2023, 17, 24154–24169. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Masuda, K.; Groso, C.; Hassan, R.; Zhou, Z.; Broderick, K.; Tsuji, M.; Tison, C. Microfluidic Synthesis of Scalable Layer-by-Layer Multiple Antigen Nano-Delivery Platform for SARS-CoV-2 Vaccines. Vaccines 2024, 12, 339. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Feng, Q.; Wang, J.; Sun, J.; Shi, X.; Jiang, X. Microfluidic synthesis of rigid nanovesicles for hydrophilic reagents delivery. Angew. Chem. Int. Ed. Engl. 2015, 54, 3952–3956. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Passos Gibson, V.; Tahiri, H.; Gilbert, C.; Yang, C.; Phan, Q.T.; Banquy, X.; Hardy, P. Modulating the Nature of Ionizable Lipids and Number of Layers in Hyaluronan-Decorated Lipid Nanoparticles for In Vitro Delivery of RNAi. Pharmaceutics 2024, 16, 563. https://doi.org/10.3390/pharmaceutics16040563
Passos Gibson V, Tahiri H, Gilbert C, Yang C, Phan QT, Banquy X, Hardy P. Modulating the Nature of Ionizable Lipids and Number of Layers in Hyaluronan-Decorated Lipid Nanoparticles for In Vitro Delivery of RNAi. Pharmaceutics. 2024; 16(4):563. https://doi.org/10.3390/pharmaceutics16040563
Chicago/Turabian StylePassos Gibson, Victor, Houda Tahiri, Claudia Gilbert, Chun Yang, Quoc Thang Phan, Xavier Banquy, and Pierre Hardy. 2024. "Modulating the Nature of Ionizable Lipids and Number of Layers in Hyaluronan-Decorated Lipid Nanoparticles for In Vitro Delivery of RNAi" Pharmaceutics 16, no. 4: 563. https://doi.org/10.3390/pharmaceutics16040563





