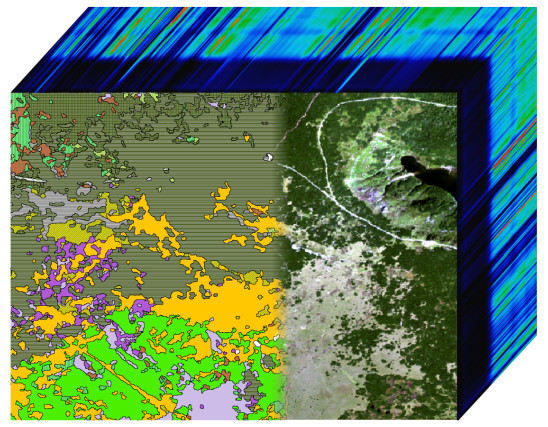
Classification of High-Mountain Vegetation Communities within a Diverse Giant Mountains Ecosystem Using Airborne APEX Hyperspectral Imagery
Abstract
:1. Introduction
- Can APEX hyperspectral image data and SVM method be used for classification of high-mountain vegetation communities in both the Polish and Czech parts of Giant Mountains?
- Does the iterative assessment of accuracy allow to distinguish communities in terms of the difficulty of identifying them?
- How does the preparation of the dataset and number of sample pixels affect the accuracy?
- What is the consistency of the classification results with the reference data?
- How can the evaluated algorithm and the results be useful for national parks administration?
2. Materials and Methods
2.1. Study Area
2.2. Reference Data
2.3. Hyperspectral Data Processing
2.4. Dimensionality Reduction
2.5. Support Vector Machine Classification
2.6. Accuracy Assesment
- Randomly select training and validation pixels without replacement from all available samples. These will create training and validation datasets. During this process, a specific number of pixels was selected for each class, as described in Table 1.
- Perform model training using the training dataset.
- Perform model accuracy assessment using the validation dataset. Calculate PA and UA for each class and OA.
- Remove all samples from training and validation dataset. Repeat step 1.
3. Results
4. Discussion
4.1. Difficulties in Classification of Mountain Vegetation Communities
5. Conclusions
- The high spatial and spectral resolution of APEX data and the use of SVM allowed to accurately classify vegetation communities with high accuracies.Iterative random selection of training and validation samples made it possible to avoid the subjectivity of a single selection of data. The most varied values were obtained for classes represented by smaller training sets, heterogeneous classes difficult to identify due to small spatial extent (less than nine square metres) and location in shaded areas (Adenostyletum alliariae, Salicetum lapponum). Higher accuracies were achieved for classes consisting of more training and validation pixels, such as mountain grasslands, Rhizocarpion alpicolae, Calamagrostion, Molinia caerulea.
- The PCA reduction allowed us to keep all the information and accelerate the classification process. Increasing the size of the training set resulted in higher OA because of a more representative dataset (the highest accuracy at 300 training pixels).
- The majority of the plant communities’ extent of the Giant Mountains was similar to the reference map from 2002. However, we underline the importance of the up-to-dateness of the data. Anthropogenic communities are subject to dynamic changes: Athyrietum distentifoli suffer damage from pest outbreaks, which can be observed within few years. Some classes were aggregated into larger ones (Artemisietea vulgaris, Pinetum mugo sudeticum, areas without vegetation) due to spectral similarities.
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Körner, C. Impact of atmospheric changes on high mountain vegetation. In Mountain Environments in Changing Climates; Routledge: London, UK, 1994; pp. 155–166. [Google Scholar] [CrossRef]
- Kozłowska, A. Strefy przejścia między układami roślinnymi-analiza wieloskalowa (na przykładzie roslinności górskiej). In Prace Geograficzne; IGiPZ PAN: Warsaw, Poland, 2008; Volume 215, pp. 1–150. [Google Scholar]
- Piękoś-Mirkowa, H.; Mirek, Z. Zbiorowiska roślinne. In Przyroda Tatrzańskiego Parku Narodowego; Mirek, Z., Ed.; Tatrzański Park Narodowy: Zakopane-Kraków, Poland, 1996; pp. 237–272. [Google Scholar]
- Müllerová, J. Use of digital aerial photography for sub-alpine vegetation mapping: A case study from the Krkonoše Mts., Czech Republic. Plant Ecol. 2005, 175, 259–272. [Google Scholar] [CrossRef]
- Jensen, J.R. Biophysical remote sensing. Ann. Assoc. Am. Geogr. 1983, 73, 111–132. [Google Scholar] [CrossRef]
- Zagajewski, B. Assessment of neural networks and Imaging Spectroscopy for vegetation classification of the High Tatras. Teledetekcja Środowiska 2010, 43, 1–113. [Google Scholar]
- Zhang, C.; Xie, Z. Object-based vegetation mapping in the Kissimmee River watershed using HyMap data and machine learning techniques. Wetlands 2013, 33, 233–244. [Google Scholar] [CrossRef]
- Burai, P.; Deák, B.; Valkó, O.; Tomor, T. Classification of herbaceous vegetation using airborne hyperspectral imagery. Remote Sens. 2015, 7, 2046–2066. [Google Scholar] [CrossRef]
- Kupková, L.; Červená, L.; Suchá, R.; Jakešová, L.; Zagajewski, B.; Březina, S.; Albrechtová, J. Classification of Tundra Vegetation in the Krkonoše Mts. National Park Using APEX, AISA Dual and Sentinel-2A Data. Eur. J. Remote Sens. 2017, 50, 29–46. [Google Scholar] [CrossRef]
- Kokaly, R.F.; Despain, D.G.; Clark, R.N.; Livo, K.E. Mapping vegetation in Yellowstone National Park using spectral feature analysis of AVIRIS data. Remote Sens. Environ. 2003, 84, 437–456. [Google Scholar] [CrossRef]
- Filippi, A.M.; Jensen, J.R. Fuzzy learning vector quantization for hyperspectral coastal vegetation classification. Remote Sens. Environ. 2006, 100, 512–530. [Google Scholar] [CrossRef]
- Jollineau, M.Y.; Howarth, P.J. Mapping an inland wetland complex using hyperspectral imagery. Int. J. Remote Sens. 2008, 29, 3609–3631. [Google Scholar] [CrossRef]
- Chan, J.; Paelinckx, D. Evaluation of Random Forest and Adaboost tree-based ensemble classification and spectral band selection for ecotope mapping using airborne hyperspectral imagery. Remote Sens. Environ. 2008, 112, 2999–3011. [Google Scholar] [CrossRef]
- Olesiuk, D.; Zagajewski, B. Wykorzystanie obrazów hiperspektralnych do klasyfikacji pokrycia terenu zlewni Bystrzanki. Teledetekcja Środowiska 2008, 40, 125–148. [Google Scholar]
- Delalieux, S.; Somers, B.; Haest, B.; Kooistra, L.; Mücher, C.A.; Vanden Borre, J. Monitoring heathland habitat status using hyperspectral image classification and unmixing. In Proceedings of the 2nd Whispers on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Iceland, 14–16 June 2010; pp. 50–54. [Google Scholar] [CrossRef]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Koch, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Neumann, C.; Förster, M.; Buddenbaum, H.; Ghosh, A.; Clasen, A.; Joshi, P.K.; Koch, B. Comparison of feature reduction algorithms for classifying tree species with hyperspectral data on three central European test sites. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2547–2561. [Google Scholar] [CrossRef]
- Tagliabue, G.; Panigada, C.; Colombo, R.; Fava, F.; Cilia, C.; Baret, F.; Vreys, K.; Meuleman, K.; Rossini, M. Forest species mapping using airborne hyperspectral APEX data. Misc. Geogr. 2016, 20, 28–33. [Google Scholar] [CrossRef]
- Raczko, E.; Zagajewski, B. Comparison of Support Vector Machine, Random Forest and Neural Network Classifiers for Tree Species Classification on Airborne Hyperspectral APEX images. Eur. J. Remote Sens. 2017, 50, 144–154. [Google Scholar] [CrossRef]
- Wang, C.; Menenti, M.; Stoll, M.P.; Belluco, E.; Marani, M. Mapping mixed vegetation communities in salt marshes using airborne spectral data. Remote Sens. Environ. 2007, 107, 559–570. [Google Scholar] [CrossRef]
- Dehaan, R.; Louis, J.; Wilson, A.; Hall, A.; Rumbachs, R. Discrimination of blackberry (Rubus fruticosus sp. agg.) using hyperspectral imagery in Kosciuszko National Park, NSW, Australia. ISPRS J. Photogramm. Remote Sens. 2007, 62, 13–24. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Gomez-Chova, L.; Calpe-Maravilla, J.; Martin-Guerrero, J.D.; Soria-Olivas, E.; Alonso-Chorda, L.; Moreno, J. Robust support vector method for hyperspectral data classification and knowledge discovery. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1530–1542. [Google Scholar] [CrossRef]
- Chan, J.C.W.; Beckers, P.; Spanhove, T.; Borre, J.V. An evaluation of ensemble classifiers for mapping Natura 2000 heathland in Belgium using spaceborne angular hyperspectral (CHRIS/Proba) imagery. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 13–22. [Google Scholar] [CrossRef]
- Pal, M.; Mather, P.M. Some issues in the classification of DAIS hyperspectral data. Int. J. Remote Sens. 2006, 27, 2895–2916. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of Hyperspectral Remote Sensing Images with Support Vector Machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Marcinkowska-Ochtyra, A.; Zagajewski, B.; Ochtyra, A.; Jarocińska, A.; Raczko, E.; Wojtuń, B. Mapping subalpine and alpine vegetation using APEX hyperspectral data. In Proceedings of the 36th EARSeL Symposium 2016, Bonn, Germany, 20–24 June 2016. [Google Scholar]
- Itten, K.I.; Dell’Endice, F.; Hueni, A.; Kneubühler, M.; Schläpfer, D.; Odermatt, D.; Seidel, D.; Huber, S.; Schopfer, J.; Kellenberger, T.; et al. APEX—The hyperspectral ESA airborne prism experiment. Sensors 2008, 8, 6235–6259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hueni, A.; Schlaepfer, D.; Jehle, M.; Schaepman, M.E. Impacts of dichroic prism coatings on radiometry of the airborne imaging spectrometer APEX. Appl. Opt. 2014, 53, 5344–5352. [Google Scholar] [CrossRef] [PubMed]
- Sterckx, S.; Vreys, K.; Biesemans, J.; Iordache, M.D.; Bertels, L.; Meuleman, K. Atmospheric correction of APEX hyperspectral data. Misc Geogr. 2016, 20, 16–20. [Google Scholar] [CrossRef]
- Vreys, K.; Iordache, M.D.; Biesemans, J.; Meuleman, K. Geometric correction of APEX hyperspectral data. Misc Geogr. 2016, 20, 11–15. [Google Scholar] [CrossRef]
- Demarchi, L.; Canters, F.; Cariou, C.; Licciardi, G.; Chan, J.C.W. Assessing the performance of two unsupervised dimensionality reduction techniques on hyperspectral APEX data for high resolution urban land-cover mapping. ISPRS J. Photogramm. Remote Sens. 2014, 87, 166–179. [Google Scholar] [CrossRef]
- Marcinkowska, A.; Zagajewski, B.; Ochtyra, A.; Jarocińska, A.; Raczko, E.; Kupková, L.; Stych, P.; Meuleman, K. Mapping vegetation communities of Karkonosze National Park using APEX hyperspectral data and Support Vector Machines. Misc. Geogr. 2014, 18, 23–29. [Google Scholar] [CrossRef]
- Jarocińska, A.; Kacprzyk, M.; Marcinkowska-Ochtyra, A.; Ochtyra, A.; Zagajewski, B.; Meuleman, K. The application of APEX images in the assessment of the state of non-forest vegetation in the Karkonosze Mountains. Misc. Geogr. 2016, 20, 21–27. [Google Scholar] [CrossRef]
- Marcinkowska-Ochtyra, A.; Zagajewski, B.; Ochtyra, A.; Jarocińska, A.; Wojtuń, B.; Rogass, C.; Mielke, C.; Lavender, S. Subalpine and Alpine Vegetation Classification based on Hyperspectral APEX and Simulated EnMAP images. Int. J. Remote Sens. 2017, 38, 1839–1864. [Google Scholar] [CrossRef]
- Suchá, R.; Jakešová, L.; Kupková, L.; Červená, L. Classification of vegetation above the tree line in the Krkonoše Mts. National Park using remote sensing multispectral data. Acta Univ. Carol. Geogr. 2016, 51, 113–129. [Google Scholar] [CrossRef]
- Żołnierz, L.; Wojtuń, B. Roślinność Subalpejska I Alpejska. In Przyroda Karkonoskiego Parku Narodowego; Knapik, R., Raj, A., Eds.; Karkonoski Park Narodowy: Jelenia Góra, Poland, 2013; pp. 241–278. [Google Scholar]
- Soukupová, L.; Kociánová, M.; Jeník, J.; Sekyra, J. Arctic-alpine tundra in the Krkonoše, the Sudetes. Opera Corcon. 1995, 32, 5–88. [Google Scholar]
- Sobik, M.; Błaś, M.; Migała, M.; Godek, M.; Nasiółkowski, T. Klimat. In Przyroda Karkonoskiego Parku Narodowego; Knapik, R., Raj, A., Eds.; Karkonoski Park Narodowy: Jelenia Góra, Poland, 2013; pp. 147–186. [Google Scholar]
- Dunajski, A. Huge dieback of the tall fern community (Athyrietum distentifolii). In Proceedings of the 9th Conference Geoecological Problems of the Krkonoše/Karkonosze Mountains: The International Scientific Conference: Past, Present and Future of Transboundary Cooperation in Research and Management, Karkonoski Park Narodowy, Szklarska Poręba, Poland, 12–14 October 2016. [Google Scholar]
- Pawłowski, B. Flora Polska; PWN: Warszawa-Kraków, Poland, 1967; Volume 11, p. 371. [Google Scholar]
- Żołnierz, L.; Wojtuń, B.; Przewoźnik, L. Ekosystemy Nieleśne Karkonoskiego Parku Narodowego; Karkonoski Park Narodowy: Jelenia Góra, Poland, 2012. [Google Scholar]
- Hejcman, M.; Češková, M.; Pavlů, V. Control of Molinia caerulea by cutting management on sub-alpine grassland. Flora Morphol. Distrib. Funct. Ecol. Plants 2010, 205, 577–582. [Google Scholar] [CrossRef]
- Matuszkiewicz, W.; Matuszkiewicz, A. Mapa zbiorowisk roślinnych Karkonoskiego Parku Narodowego. Ochr. Przyr. 1974, 40, 45–109. [Google Scholar]
- Danielewicz, W.; Raj, A.; Zientarski, J. Lasy. In Przyroda Karkonoskiego Parku Narodowego; Knapik, R., Raj, A., Eds.; Karkonoski Park Narodowy: Jelenia Góra, Poland, 2013; pp. 241–278. [Google Scholar]
- Wojtuń, B.; Żołnierz, L. Plan Ochrony Ekosystemów Nieleśnych—Inwentaryzacja Zbiorowisk. In Plan Ochrony Karkonoskiego Parku Narodowego; Bureau for Forest Management and Geodesy: Brzeg, Poland, 2002. [Google Scholar]
- Itten, K.I.; Schaepman, M.; De Vos, L.; Hermans, L.; Schlaepfer, H.; Space, O.C.; Droz, F. APEX–airborne prism experiment a new concept for an airborne imaging spectrometer. In Proceedings of the Third International Airborne Remote Sensing Conference and Exhibition, Copenhagen, Denmark, 7–10 July 1997; Volume 1, pp. 181–188. [Google Scholar]
- Zhu, Z.; Woodcock, C.E. Object-based cloud and cloud shadow detection in Landsat imagery. Remote Sens. Environ. 2012, 118, 83–94. [Google Scholar] [CrossRef]
- Hotelling, H. Analysis of a complex of statistical variables into principal components. J. Educat. Psychol. 1933, 24, 417–441. [Google Scholar] [CrossRef]
- Green, A.A.; Berman, M.; Switzer, P.; Craig, M.D. A transformation for ordering multispectral data in terms of image quality with implications for noise removal. IEEE Trans. Geosci. Remote Sens. 1988, 26, 65–74. [Google Scholar] [CrossRef]
- John, G.H.; Kohavi, R.; Pfleger, K. Irrelevant features and the subset selection problem. In Proceedings of the Eleventh International Machine Learning Conference, Rutgers University, New Brunswick, NJ, USA, 10–13 July 1994; pp. 121–129. [Google Scholar] [CrossRef]
- Vapnik, V.N. The Nature of Statistical Learning Theory; Springer: New York, NY, USA, 1995. [Google Scholar] [CrossRef]
- Gualtieri, J.A.; Cromp, R.F. Support vector machines for hyperspectral remote sensing classification. In Proceedings of the 27th AIPR Workshop: Advances in Computer Assisted Recognition; SPIE: Bellingham, WA, USA, 1999; pp. 221–232. [Google Scholar] [CrossRef]
- Hsu, C.W.; Chang, C.C.; Lin, C.J. A Practical Guide to Support Vector Classification; National Taiwan University, 2010; Available online: https://ntu.csie.org/~cjlin/papers/guide/guide.pdf (accessed on 13 February 2017).
- Liao, R. Support Vector Machines, CSC 411 Tutorial. 2015. Available online: https://.cs.toronto.edu/~urtasun/courses/CSC411/tutorial9.pdf (accessed on 5 September 2016).
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F. e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien. R Package Version 1.6-8. 2017. Available online: https://CRAN.R-project.org/package=e1071 (accessed on 15 December 2017).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https://www.R-project.org/ (accessed on 15 January 2018).
- Congalton, R.G. A review of assessing the accuracy of classifications of remotely sensed data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Marcinkowska-Ochtyra, A. Assessment of APEX Hyperspectral Images and Support Vector Machines for Karkonosze Subalpine and Alpine Vegetation Classification. Ph.D. Thesis, University of Warsaw, Warsaw, Poland, 28 November 2016. [Google Scholar]
- Raczko, E.; Zagajewski, B.; Ochtyra, A.; Jarocińska, A.; Marcinkowska-Ochtyra, A.; Dobrowolski, M. Forest species identification of Mount Chojnik (Karkonoski National Park) using airborne hyperspectal APEX data. Sylwan 2015, 159, 593–599. [Google Scholar]
- Foody, G.M.; Mathur, A. A relative evaluation of multi-class image classification by support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1335–1343. [Google Scholar] [CrossRef]
- Lin, S.W.; Lee, Z.J.; Chen, S.C.; Tseng, T.Y. Parameter determination of support vector machine and feature selection using simulated annealing approach. Appl. Soft Comput. 2008, 8, 1505–1512. [Google Scholar] [CrossRef]
- Waske, B.; van der Linden, S.; Benediktsson, J.A.; Rabe, A.; Hostert, P. Sensitivity of Support Vector Machines to Random Feature Selection in Classification of Hyperspectral Data. IEEE Trans. Geosci. Remote Sens. 2010, 48, 2880–2889. [Google Scholar] [CrossRef]
- Kozłowska, A. Problemy kartowania roślinności wysokogórskiej w skali szczegółowej (na przykładzie map roślinności Kotła Gąsienicowego i Goryczkowego Świńskiego). In Badania Geoekologiczne w Otoczeniu Kasprowego Wierchu; Kotarba, A., Kozłowska, A., Eds.; Prace Geograficzne, Volume 174; Wydawnictwo Continuo: Wrocław, Poland, 1999; pp. 37–43. [Google Scholar]






| No. | Class | Training Samples | Validation Samples | ||
|---|---|---|---|---|---|
| Original | Randomly Selected | Original | Randomly Selected | ||
| 1 | Rhizocarpion alpicolae all. 1 | 582 | 200 | 1116 | 400 |
| 2 | Umbilicarion cylindricae all. | 170 | 150 | 341 | 300 |
| 3 | Carici (rigidae)–Nardetum | 546 | 200 | 1079 | 400 |
| 4 | Carici (rigidae)–Festucetum airoidis (subalpine) | 267 | 200 | 389 | 300 |
| 5 | Carici (rigidae)–Festucetum airoidis (alpine) | 116 | 100 | 235 | 200 |
| 6 | Cardamino–Montion all. | 30 | 30 | 62 | 60 |
| 7 | Scheuchzerio–Caricetea nigrae cl. 2 | 236 | 200 | 503 | 400 |
| 8 | Oxycocco–Sphagnetea nigrae cl. | 345 | 200 | 509 | 400 |
| 9 | Crepido–Calamagrostietum villosae | 456 | 200 | 668 | 400 |
| 10 | Deschampsia caespitosa | 496 | 200 | 1005 | 400 |
| 11 | Molinia caerulea | 347 | 200 | 525 | 400 |
| 12 | Calamagrostion all. | 331 | 200 | 593 | 400 |
| 13 | Pado–Sorbetum | 193 | 200 | 440 | 400 |
| 14 | Salicetum lapponum | 58 | 50 | 111 | 100 |
| 15 | Adenostyletum alliariae | 12 | 10 | 25 | 20 |
| 16 | Athyrietum distentifolii | 503 | 200 | 1108 | 400 |
| 17 | Empetro–Vaccinietum | 159 | 150 | 322 | 300 |
| 18 | Vaccinium myrtillus | 668 | 200 | 1351 | 400 |
| 19 | Calluna vulgaris | 67 | 50 | 128 | 100 |
| 20 | Artemisietea vulgaris cl. | 39 | 30 | 80 | 60 |
| 21 | Pinetum mugo sudeticum | 2463 | 200 | 5501 | 400 |
| 22 | Calamagrostio villosae–Piceetum | 1000 | 200 | 2002 | 400 |
| 23 | lakes | 524 | 200 | 1076 | 400 |
| 24 | areas without vegetation | 239 | 200 | 471 | 400 |
| Dataset | Overall accuracy (OA, %) | File Size (MB) | |
|---|---|---|---|
| Linear | Radial | ||
| 252 spectral bands | 82.69 | 83.11 | 465 |
| 40 PCA bands | 81.04 | 84.49 | 65 |
| 30 MNF bands | 80.76 | 82.02 | 48 |
| 70 spectral bands * | 76.68 | 77.16 | 113 |
| 18 spectral bands * | 68.14 | 69.01 | 31 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcinkowska-Ochtyra, A.; Zagajewski, B.; Raczko, E.; Ochtyra, A.; Jarocińska, A. Classification of High-Mountain Vegetation Communities within a Diverse Giant Mountains Ecosystem Using Airborne APEX Hyperspectral Imagery. Remote Sens. 2018, 10, 570. https://doi.org/10.3390/rs10040570
Marcinkowska-Ochtyra A, Zagajewski B, Raczko E, Ochtyra A, Jarocińska A. Classification of High-Mountain Vegetation Communities within a Diverse Giant Mountains Ecosystem Using Airborne APEX Hyperspectral Imagery. Remote Sensing. 2018; 10(4):570. https://doi.org/10.3390/rs10040570
Chicago/Turabian StyleMarcinkowska-Ochtyra, Adriana, Bogdan Zagajewski, Edwin Raczko, Adrian Ochtyra, and Anna Jarocińska. 2018. "Classification of High-Mountain Vegetation Communities within a Diverse Giant Mountains Ecosystem Using Airborne APEX Hyperspectral Imagery" Remote Sensing 10, no. 4: 570. https://doi.org/10.3390/rs10040570