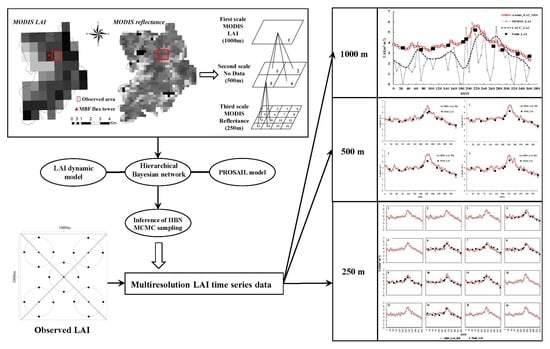
Assimilating Multiresolution Leaf Area Index of Moso Bamboo Forest from MODIS Time Series Data Based on a Hierarchical Bayesian Network Algorithm
Abstract
:1. Introduction
2. Study Area and Datasets
2.1. Study Area
2.2. Datasets and Processing
2.2.1. Processing of Satellite Data
2.2.2. Processing of Observed Data
3. Study Methodology
- The 1000-m-resolution MODIS LAI and the 250-m-resolution MODIS reflectance data were processed, which had been carried out in Section 2.2. Smoothed MODIS LAI data were input into the LAI dynamic model to obtain the simulated LAI.
- Based on the 250-m-resolution MODIS reflectance data, resolution-specific likelihood inference (RESL) or resolution-specific restricted-likelihood inference (RESREL) method was used to estimate the parameters in the data assimilation.
- The LAI data and simulated reflectance data at each scale were obtained using the simulated LAI, parameters in the process model of the HBN, and the PROSAIL model. Combined with MODIS reflectance data, the structure of the HBN at three resolutions (1000, 500, and 250 m) was initialized.
- An inference of HBN was used to update node (pixel) states by integrating observations with different spatial resolutions. The probability distributions of node states were updated by upward filtering from observations from the finer scale (250 m) to the coarser scale (1000 m). Then, the downward smoothing started from the coarser scale (1000 m) and ended at the finer scale (250 m) to update the posterior probability distributions of all node states according to all of the observations.
- The Markov chain Monte Carlo (MCMC) sampling method was used to sample the probability distribution. The LAI corresponding to the maximum posterior probability was the assimilation result of the three scales.
- Because of uncertainty and error in the parameter estimation of the HBN, the above process was repeated 100 times and the average value was used as the final assimilation LAI. The assimilated LAI time series data were compared with the observed LAI. The methodology of this HBN algorithm was implemented within MATLAB.
3.1. LAI Dynamic Model
3.2. PROSAIL Model
3.3. Hierarchical Bayesian Network Algorithm
3.3.1. Construction of the HBN
3.3.2. Inference of the HBN
3.3.3. MCMC Sampling
3.4. Accuracy Assessment
4. Results and Analysis
4.1. Validation of LAI Assimilation at 1000-m Resolution
4.2. Validation of LAI Assimilation at 500-m Resolution
4.3. Validation of LAI Assimilation at 250-m Resolution
5. Discussion and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, J.; Black, T.A. Defining leaf area index for non-flat leaves. Plant Cell Environ. 1992, 15, 421–429. [Google Scholar] [CrossRef]
- Sellers, P.J.; Dickinson, R.E.; Randall, D.A.; Betts, A.K.; Hall, F.G.; Berry, J.A.; Collatz, G.J.; Denning, A.S.; Mooney, H.A.; Nobre, C.A. Modeling the Exchanges of Energy, Water, and Carbon between Continents and the Atmosphere. Science 1997, 275, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Pasolli, L.; Asam, S.; Castelli, M.; Bruzzone, L.; Wohlfahrt, G.; Zebisch, M.; Notarnicola, C. Retrieval of Leaf Area Index in mountain grasslands in the Alps from MODIS satellite imagery. Remote Sens. Environ. 2015, 165, 159–174. [Google Scholar] [CrossRef]
- Kauwe, M.G.D.; Disney, M.I.; Quaife, T.; Lewis, P.; Williams, M. An assessment of the MODIS collection 5 leaf area index product for a region of mixed coniferous forest. Remote Sens. Environ. 2011, 115, 767–780. [Google Scholar] [CrossRef]
- Jonckheere, I.; Fleck, S.; Nackaerts, K.; Muys, B.; Coppin, P.; Weiss, M.; Baret, F. Review of methods for in situ leaf area index determination: Part I. Theories, sensors and hemispherical photography. Agric. For. Meteorol. 2004, 121, 19–35. [Google Scholar] [CrossRef]
- Dong, T.; Liu, J.; Qian, B.; Zhao, T.; Jing, Q.; Geng, X.; Wang, J.; Huffman, T.; Shang, J. Estimating winter wheat biomass by assimilating leaf area index derived from fusion of Landsat-8 and MODIS data. Int. J. Appl. Earth Obs. Geoinf. 2016, 49, 63–74. [Google Scholar] [CrossRef]
- Li, X.; Xiao, Z.; Wang, J.; Ying, Q.U.; Jin, H. Dual Ensemble Kalman Filter assimilation method for estimating time series LAI. J. Remote Sens. 2014, 18, 27–44. [Google Scholar] [CrossRef]
- Gray, J.; Song, C. Mapping leaf area index using spatial, spectral, and temporal information from multiple sensors. Remote Sens. Environ. 2012, 119, 173–183. [Google Scholar] [CrossRef]
- Myneni, R.B.; Hoffman, S.; Knyazikhin, Y.; Privette, J.L.; Glassy, J.; Tian, Y.; Wang, Y.; Song, X.; Zhang, Y.; Smith, G.R. Global products of vegetation leaf area and fraction absorbed PAR from year one of MODIS data. Remote Sens. Environ. 2002, 83, 214–231. [Google Scholar] [CrossRef] [Green Version]
- Baret, F.; Hagolle, O.; Geiger, B.; Bicheron, P.; Miras, B.; Huc, M.; Berthelot, B.; Niño, F.; Weiss, M.; Samain, O. LAI, fAPAR and fCover CYCLOPES global products derived from VEGETATION: Part 1: Principles of the algorithm. Remote Sens. Environ. 2007, 110, 275–286. [Google Scholar] [CrossRef]
- Deng, F.; Chen, J.M.; Plummer, S.; Chen, M.; Pisek, J. Algorithm for global leaf area index retrieval using satellite imagery. IEEE Trans. Geosci. Remote Sens. 2006, 44, 2219–2229. [Google Scholar] [CrossRef] [Green Version]
- Masson, V.; Champeaux, J.L.; Chauvin, F.; Meriguet, C.; Lacaze, R. A Global Database of Land Surface Parameters at 1-km Resolution in Meteorological and Climate Models. J. Clim. 2003, 16, 1231–1281. [Google Scholar] [CrossRef]
- Gao, F.; Morisette, J.T.; Wolfe, R.E.; Ederer, G.; Pedelty, J.; Masuoka, E.; Myneni, R.; Tan, B.; Nightingale, J. An Algorithm to Produce Temporally and Spatially Continuous MODIS-LAI Time Series. IEEE Geosci. Remote Sens. Lett. 2008, 5, 60–64. [Google Scholar] [CrossRef]
- Fang, H.; Liang, S.; Townshend, J.R.; Dickinson, R.E. Spatially and temporally continuous LAI data sets based on an integrated filtering method: Examples from North America. Remote Sens. Environ. 2013, 112, 75–93. [Google Scholar] [CrossRef]
- Borak, J.S.; Jasinski, M.F. Effective interpolation of incomplete satellite-derived leaf-area index time series for the continental United States. Agric. For. Meteorol. 2008, 149, 320–332. [Google Scholar] [CrossRef]
- Ma, J.; Qin, S. Recent Advances and Development of Data Assimilation Algorithms. Adv. Earth Sci. 2012, 27, 747–757. [Google Scholar] [CrossRef]
- Mclaughlin, D.; Miller, C.T.; Parlange, M.B.; Hassanizadeh, S.M. An integrated approach to hydrologic data assimilation: Interpolation, smoothing, and filtering. Adv. Water Resour. 2002, 25, 1275–1286. [Google Scholar] [CrossRef]
- Li, X.; Mao, F.; Du, H.; Zhou, G.; Xu, X.; Han, N.; Sun, S.; Gao, G.; Chen, L. Assimilating leaf area index of three typical types of subtropical forest in China from MODIS time series data based on the integrated ensemble Kalman filter and PROSAIL model. ISPRS J. Photogramm. Remote Sens. 2017, 126, 68–78. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, S.; Shen, S. Assimilating remote sensing information with crop model using Ensemble Kalman Filter for improving LAI monitoring and yield estimation. Ecol. Model. 2013, 270, 30–42. [Google Scholar] [CrossRef]
- Mao, F.; Li, X.; Du, H.; Zhou, G.; Han, N.; Xu, X.; Liu, Y.; Chen, L.; Cui, L. Comparison of Two Data Assimilation Methods for Improving MODIS LAI Time Series for Bamboo Forests. Remote Sens. 2017, 9, 401. [Google Scholar] [CrossRef]
- Li, H.; Chen, Z.; Wu, W.; Jiang, Z.; Liu, B.; Hasi, T. Crop model data assimilation with particle filter for yield prediction using leaf area index of different temporal scales. In Proceedings of the Fourth International Conference on Agro-Geoinformatics, Istanbul, Turkey, 20–24 July 2015; pp. 401–406. [Google Scholar]
- Li, X.; Du, H.; Mao, F.; Zhou, G.; Chen, L.; Xing, L.; Fan, W.; Xu, X.; Liu, Y.; Cui, L. Estimating bamboo forest aboveground biomass using EnKF-assimilated MODIS LAI spatiotemporal data and machine learning algorithms. Agric. For. Meteorol. 2018, 256–257, 445–457. [Google Scholar] [CrossRef]
- Li, X.; Mao, F.; Du, H.; Zhou, G.; Xu, X.; Li, P.; Liu, Y.; Cui, L. Simulating of carbon fluxes in bamboo forest ecosystem using BEPS model based on the LAI assimilated with Dual Ensemble Kalman Filter. Chin. J. Appl. Ecol. 2016. [Google Scholar] [CrossRef]
- Li, X.; Lu, H.; Yu, L.; Yang, K. Comparison of the Spatial Characteristics of Four Remotely Sensed Leaf Area Index Products over China: Direct Validation and Relative Uncertainties. Remote Sens. 2018, 10, 148. [Google Scholar] [CrossRef]
- Yuan, H.; Dai, Y.; Xiao, Z.; Ji, D.; Shangguan, W. Reprocessing the MODIS Leaf Area Index products for land surface and climate modelling. Remote Sens. Environ. 2011, 115, 1171–1187. [Google Scholar] [CrossRef]
- Jinsheng, H. Carbon cycling of Chinese forests: From carbon storage, dynamics to models. Sci. China Life Sci. 2012, 55, 188–190. [Google Scholar] [CrossRef] [Green Version]
- Cao, M.; Yu, G.; Liu, J.; Li, K. Multi-scale observation and cross-scale mechanistic modeling on terrestrial ecosystem carbon cycle. Sci. China 2005, 48, 17–32. [Google Scholar] [CrossRef]
- Xiao, Z.; Wang, J.; Wan, H. Multiscale approach for fusing leaf area index estimates from multiple sensors. Proc. SPIE 2007, 6790, 679013. [Google Scholar] [CrossRef]
- Wang, D.; Liang, S. Using multiresolution tree to integrate MODIS and MISR-L3 LAI products. In Proceedings of the IEEE International Geoscience & Remote Sensing Symposium, Honolulu, HI, USA, 25–30 July 2010; Volume 38, pp. 1027–1030. [Google Scholar] [CrossRef]
- Jiang, J.; Xiao, Z.; Wang, J.; Song, J. Multiscale Estimation of Leaf Area Index from Satellite Observations Based on an Ensemble Multiscale Filter. Remote Sens. 2016, 8, 229. [Google Scholar] [CrossRef]
- Smith, A.F.M.; Berliner, L.M.; Royle, J.A.; Wikle, C.K.; Milliff, R.F. Bayesian Methods in the Atmospheric Sciences; Oxford University Press: Oxford, UK, 1998. [Google Scholar]
- Wikle, C.K.; Anderson, C.J. Climatological analysis of tornado report counts using a hierarchical Bayesian spatiotemporal model. J. Geophys. Res. Atmos. 2003, 108. [Google Scholar] [CrossRef] [Green Version]
- Wikle, C.K.; Berliner, L.M.; Cressie, N. Hierarchical Bayesian space-time models. Environ. Ecol. Stat. 1998, 5, 117–154. [Google Scholar] [CrossRef]
- Berliner, L.M. Hierarchical Bayesian Time Series Models; Springer: Dordrecht, The Netherlands, 1996; pp. 15–22. [Google Scholar]
- Wikle, C.K.; Berliner, M.L. Combining Information Across Spatial Scales. Technometrics 2005, 47, 80–91. [Google Scholar] [CrossRef]
- Kolaczyk, E.D.; Huang, H. Multiscale Statistical Models for Hierarchical Spatial Aggregation. Geogr. Anal. 2010, 33, 95–118. [Google Scholar] [CrossRef] [Green Version]
- Berrocal, V.J.; Gelfand, A.E.; Holland, D.M. A Spatio-Temporal Downscaler for Output From Numerical Models. J. Agric. Biol. Environ. Stat. 2010, 15, 176–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahu, S.K.; Yip, S.; Holland, D.M. Improved space–time forecasting of next day ozone concentrations in the eastern US. Atmos. Environ. 2009, 43, 494–501. [Google Scholar] [CrossRef]
- Sahu, S.K.; Gelfand, A.E.; Holland, D.M. High Resolution Space-Time Ozone Modeling for Assessing Trends. J. Am. Stat. Assoc. 2007, 102, 1221. [Google Scholar] [CrossRef] [PubMed]
- Berliner, L.M.; Milliff, R.F.; Wikle, C.K. Bayesian hierarchical modeling of air-sea interaction. J. Geophys. Res. Oceans 2003, 108, 303–307. [Google Scholar] [CrossRef]
- Mcmillan, N.J.; Holland, D.M.; Morara, M.; Feng, J. Combining numerical model output and particulate data using Bayesian space-time modeling. Environmetrics 2010, 21, 48–65. [Google Scholar] [CrossRef]
- Fuentes, M.; Raftery, A.E. Model evaluation and spatial interpolation by Bayesian combination of observations with outputs from numerical models. Biometrics 2005, 61, 36. [Google Scholar] [CrossRef]
- Cocchi, D.; Greco, F.; Trivisano, C. Hierarchical space-time modelling of PM pollution. Atmos. Environ. 2007, 41, 532–542. [Google Scholar] [CrossRef]
- Gelfand, A.E.; Sahu, S.K. Combining monitoring data and computer model output in assessing environmental exposure. In Oxford Handbook of Applied Bayesian Analysis; Oxford University Press: Oxford, UK, 2010; pp. 482–510. [Google Scholar]
- Qin, S.; Ma, J.; Wang, X. Construction and Experiment of Hierarchical Bayesian Network in Data Assimilation. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 6, 1036–1047. [Google Scholar] [CrossRef]
- Qin, S.; Ma, J.; Wang, X. Development of a hierarchical Bayesian network algorithm for land surface data assimilation. Int. J. Remote Sens. 2013, 34, 1905–1927. [Google Scholar] [CrossRef]
- Du, H.; Zhou, G.; Xu, X. Quantitative Methods Using Remote Sensing in Estimating Biomass and Carbon Storage Bamboo Forest; Science Press: Beijing, China, 2012. [Google Scholar]
- Zhou, G.; Jiang, P.; Du, H.; Shi, Y. Technology for the Measurement and Enhancement Carbon Sinks in Bamboo Forest Ecosystems; Science Press: Beijing, China, 2017. [Google Scholar]
- Han, N.; Du, H.; Zhou, G.; Xu, X.; Cui, R.; Gu, C. Spatiotemporal heterogeneity of Moso bamboo aboveground carbon storage with Landsat Thematic Mapper images: A case study from Anji County, China. Int. J. Remote Sens. 2013, 34, 4917–4932. [Google Scholar] [CrossRef]
- Du, H.; Zhou, G.; Ge, H.; Fan, W.; Xu, X.; Fan, W.; Shi, Y. Satellite-based carbon stock estimation for bamboo forest with a non-linear partial least square regression technique. Int. J. Remote Sens. 2012, 33, 1917–1933. [Google Scholar] [CrossRef]
- Zhou, G.; Xu, X.; Du, H.; Ge, H.; Shi, Y.; Zhou, Y. Estimating Aboveground Carbon of Moso Bamboo Forests Using the k Nearest Neighbors Technique and Satellite Imagery. Photogramm. Eng. Remote Sens. 2011, 77, 1123–1131. [Google Scholar] [CrossRef]
- Chen, J.M.; Deng, F.; Chen, M. Locally adjusted cubic-spline capping for reconstructing seasonal trajectories of a satellite-derived surface parameter. IEEE Trans. Geosci. Remote Sens. 2006, 44, 2230–2238. [Google Scholar] [CrossRef]
- Xiao, Z.; Liang, S.; Wang, J.; Jiang, B.; Li, X. Real-time retrieval of Leaf Area Index from MODIS time series data. Remote Sens. Environ. 2011, 115, 97–106. [Google Scholar] [CrossRef]
- Li, X. Assimilation of MODIS LAI Time Series in Bamboo Forest and Its Application in Carbon Flux Simulation; Zhejiang A&F University: Hangzhou, China, 2017. [Google Scholar]
- Sun, S.; Du, H.; Li, P.; Zhou, G.; Xu, X.; Gao, G.; Li, X. Retrieval of leaf net photosynthetic rate of moso bamboo forests using hyperspectral remote sen-sing based on wavelet transform. Chin. J. Appl. Ecol. 2016, 27, 49–58. [Google Scholar] [CrossRef]
- Lu, G.; Du, H.; Zhou, G.; Lv, Y.; Gu, C.; Shang, Z. Dynamic change of Phyllostachys edulis forest canopy parameters and their relationships with photosynthetic active radiation in the bamboo shooting growth phase. J. Zhejiang A F Univ. 2012, 29, 844–850. [Google Scholar] [CrossRef]
- Dickinson, R.E.; Tian, Y.; Liu, Q.; Zhou, L. Dynamics of leaf area for climate and weather models. J. Geophys. Res. Atmos. 2008, 113. [Google Scholar] [CrossRef] [Green Version]
- Jacquemoud, S.; Verhoef, W.; Baret, F.; Bacour, C.; Zarcotejada, P.J.; Asner, G.P.; François, C.; Ustin, S.L.; Ustin, S.L.; Schaepman, M.E. PROSPECT+SAIL models: A review of use for vegetation characterization. Remote Sens. Environ. 2009, 113, S56–S66. [Google Scholar] [CrossRef]
- Feret, J.B.; François, C.; Asner, G.P.; Gitelson, A.A.; Martin, R.E.; Bidel, L.P.R.; Ustin, S.L.; Maire, G.L.; Jacquemoud, S. PROSPECT-4 and 5: Advances in the leaf optical properties model separating photosynthetic pigments. Remote Sens. Environ. 2008, 112, 3030–3043. [Google Scholar] [CrossRef]
- Verhoef, W.; Bach, H. Coupled soil–leaf-canopy and atmosphere radiative transfer modeling to simulate hyperspectral multi-angular surface reflectance and TOA radiance data. Remote Sens. Environ. 2007, 109, 166–182. [Google Scholar] [CrossRef]
- Gu, C.; Du, H.; Zhou, G.; Han, N.; Xu, X.; Zhao, X.; Sun, X. Retrieval of leaf area index of moso bamboo forest with Landsat Thematic Mapper image based on PROSAIL canopy radiative transfer model. Chin. J. Appl. Ecol. 2013, 24, 2248–2256. [Google Scholar] [CrossRef]
- Wikle, C.K.; Berliner, L.M. A Bayesian tutorial for data assimilation. Phys. D Nonlinear Phenom. 2007, 230, 1–16. [Google Scholar] [CrossRef]
- Ma, J. Data Assimilation Algorithm Development and Experiment; Science Press: Beijing, China, 2013. [Google Scholar]
- Gybels, J.; Martin, P. Multi-Resolution Statistical Modeling in Space and Time with Application to Remote Sensing of the Environment; Ohio State University: Columbus, OH, USA, 2003. [Google Scholar]
- Harville, D.A. Matrix Algebra From a Statistician’s Perspective; Springer: New York, NY, USA, 1997. [Google Scholar]
- Mcculloch, C.E.; Searle, S.R. Generalized, Linear, and Mixed Models; Wiley: Hoboken, NJ, USA, 2008. [Google Scholar]
- Ferreira, M.A.R.; Lee, H.K.H. Multiscale Modeling: A Bayesian Perspective; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Huang, H.C.; Cressie, N.; Gabrosek, J. Fast, Resolution-Consistent Spatial Prediction of Global Processes From Satellite Data. J. Comput. Graph. Stat. 2002. [Google Scholar] [CrossRef]
- Geweke, J. Bayesian Inference in Econometric Models Using Monte Carlo Integration. Econometrica 1989, 57, 1317–1339. [Google Scholar] [CrossRef]
- Hastings, W.K. Monte Carlo Sampling Methods Using Markov Chains and Their Applications. Biometrika 1970, 57, 97–109. [Google Scholar] [CrossRef]
- Smith, A.F.M.; Roberts, G.O. Bayesian Computation Via the Gibbs Sampler and Related Markov Chain Monte Carlo Methods. J. R. Stat. Soc. 1993, 55, 3–23. [Google Scholar] [CrossRef]
- Flötteröd, G.; Bierlaire, M. Metropolis–Hastings sampling of paths. Transp. Res. Part B 2013, 48, 53–66. [Google Scholar] [CrossRef]
- Geweke, J.; Tanizaki, H. Bayesian estimation of state-space models using the Metropolis–Hastings algorithm within Gibbs sampling. Comput. Stat. Data Anal. 2001, 37, 151–170. [Google Scholar] [CrossRef] [Green Version]
- Chib, S.; Greenberg, E. Understanding the Metropolis-Hastings Algorithm. Am. Stat. 1995, 49, 327–335. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Tan, B.; Huang, D.; Rautiainen, M.; Shabanov, N.V.; Wang, Y.; Privette, J.L.; Huemmrich, K.F.; Fensholt, R.; Sandholt, I. MODIS leaf area index products: From validation to algorithm improvement. IEEE Trans. Geosci. Remote Sens. 2006, 44, 1885–1898. [Google Scholar] [CrossRef]











| DOY | 23 | 70 | 103 | 142 | 181 | 195 | 217 | 263 | 290 | 330 | 363 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LAI | ||||||||||||
| LAI_1000 | 3.51 | 3.33 | 3.43 | 3.76 | 4.08 | 4.35 | 5.22 | 3.67 | 3.31 | 2.94 | 2.72 | |
| LAI_500_1 | 3.74 | 3.39 | 3.61 | 3.79 | 4.13 | 4.47 | 4.99 | 3.94 | 3.81 | 2.96 | 2.76 | |
| LAI_500_2 | 3.71 | 3.53 | 3.98 | 3.71 | 3.90 | 4.45 | 5.29 | 3.82 | 3.48 | 3.38 | 2.70 | |
| LAI_500_3 | 3.72 | 3.55 | 3.67 | 3.82 | 4.09 | 4.34 | 5.16 | 3.98 | 3.49 | 3.15 | 2.79 | |
| LAI_500_4 | 3.97 | 3.48 | 3.84 | 3.45 | 3.92 | 4.19 | 5.30 | 3.70 | 3.58 | 3.00 | 2.77 | |
| LAI_250_4 | 3.74 | 3.39 | 3.61 | 3.79 | 4.13 | 4.47 | 4.99 | 3.94 | 3.81 | 2.96 | 2.76 | |
| LAI_250_6 | 3.86 | 4.00 | 4.14 | 3.85 | 4.54 | 4.92 | 5.37 | 4.33 | 3.87 | 3.73 | 2.82 | |
| LAI_250_7 | 3.85 | 3.78 | 3.98 | 3.72 | 4.44 | 4.58 | 5.21 | 4.54 | 4.33 | 3.85 | 2.89 | |
| LAI_250_8 | 3.97 | 3.53 | 4.05 | 3.57 | 3.90 | 4.21 | 5.09 | 3.82 | 3.48 | 2.91 | 2.38 | |
| LAI_250_9 | 3.68 | 3.52 | 3.89 | 4.07 | 4.16 | 4.62 | 5.05 | 4.12 | 3.42 | 3.28 | 2.89 | |
| LAI_250_10 | 3.75 | 3.66 | 3.60 | 3.61 | 4.06 | 4.33 | 5.01 | 3.92 | 3.52 | 3.08 | 2.86 | |
| LAI_250_11 | 3.59 | 3.42 | 3.91 | 4.05 | 4.08 | 4.96 | 5.25 | 4.44 | 3.55 | 3.29 | 2.09 | |
| LAI_250_14 | 3.97 | 3.48 | 3.84 | 3.45 | 3.92 | 4.19 | 5.30 | 3.70 | 3.58 | 3.00 | 2.77 | |
| Model | Parameter (Unit) | Value |
|---|---|---|
| PROSPECT5 | Leaf mesophyll structure | 1.04 |
| Chlorophyll content | 28–55 | |
| Carotenoids content | 10 | |
| Water content | 0.0035 | |
| Dry matter | 0.003 | |
| 4SAIL | Leaf area index | 0.0–7.0 |
| Average leaf angle (°) | 20.20 | |
| Hot-spot parameter | 0.0003 | |
| View zenith angle (°) | 0–90 | |
| Solar zenith angle (°) | 0–90 | |
| Relative azimuth angle (°) | 0–180 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, L.; Li, X.; Du, H.; Zhou, G.; Mao, F.; Liu, T.; Zheng, J.; Dong, L.; Zhang, M.; Han, N.; et al. Assimilating Multiresolution Leaf Area Index of Moso Bamboo Forest from MODIS Time Series Data Based on a Hierarchical Bayesian Network Algorithm. Remote Sens. 2019, 11, 56. https://doi.org/10.3390/rs11010056
Xing L, Li X, Du H, Zhou G, Mao F, Liu T, Zheng J, Dong L, Zhang M, Han N, et al. Assimilating Multiresolution Leaf Area Index of Moso Bamboo Forest from MODIS Time Series Data Based on a Hierarchical Bayesian Network Algorithm. Remote Sensing. 2019; 11(1):56. https://doi.org/10.3390/rs11010056
Chicago/Turabian StyleXing, Luqi, Xuejian Li, Huaqiang Du, Guomo Zhou, Fangjie Mao, Tengyan Liu, Junlong Zheng, Luofan Dong, Meng Zhang, Ning Han, and et al. 2019. "Assimilating Multiresolution Leaf Area Index of Moso Bamboo Forest from MODIS Time Series Data Based on a Hierarchical Bayesian Network Algorithm" Remote Sensing 11, no. 1: 56. https://doi.org/10.3390/rs11010056