Forest Stand Species Mapping Using the Sentinel-2 Time Series
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Data Collection and Preprocessing
2.3. Methods
2.3.1. Forest Mask
2.3.2. Training and Validation Samples
2.3.3. Variable Importance and Assessment of Temporal Patterns
2.3.4. Forest Tree Species Classification
2.3.5. Accuracy Assessment
3. Results
3.1. Variable Importance and Temporal Patterns
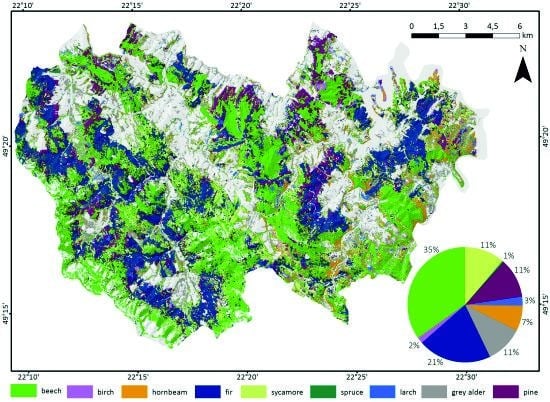
3.2. Forest Tree Species Classification
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A










References
- Ballanti, L.; Blesius, L.; Hines, E.; Kruse, B. Tree species classification using hyperspectral imagery: A comparison of two classifiers. Remote Sens. 2016, 8, 445. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Sheeren, D.; Fauvel, M.; Josipović, V.; Lopes, M.; Planque, C.; Willm, J.; Dejoux, J.-F. Tree Species Classification in Temperate Forests Using Formosat-2 Satellite Image Time Series. Remote Sens. 2016, 8, 734. [Google Scholar] [CrossRef]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Kochb, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Sedliak, M.; Sačkov, I.; Kulla, L. Classification of tree species composition using a combination of multispectral imagery and airborne laser scanning data. Cent. Eur. For. J. 2017, 63, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Mickelson, J.G.; Civco, D.L.; Silander, J.A. Delineating Forest Canopy Species in the Northeastern United States Using Multi-Temporal TM Imagery. Photogramm. Eng. Remote Sens. 1998, 64, 891–904. [Google Scholar]
- Schmitt, U.; Ruppert, G.S. Forest Classification of Multitemporal Mosaicked Satellite Images. Int. Arch. Photogramm. Remote Sens. 1996, 31, 602–605. [Google Scholar]
- Walsh, S.J. Coniferous Tree Species Mapping Using LANDSAT Data. Remote Sens. Environ. 1980, 9, 11–26. [Google Scholar] [CrossRef]
- Madonsela, S.; Cho, M.A.; Mathieu, R.; Mutanga, O.; Ramoelo, A.; Kaszta, Ż.; Van De Kerchove, R.; Wolff, E. Multi-phenology WorldView-2 imagery improves remote sensing of savannah tree species. Int. J. Appl. Earth Obs. Geoinf. 2017, 58, 65–73. [Google Scholar] [CrossRef]
- Xie, Y.; Sha, Z.; Yu, M. Remote sensing imagery in vegetation mapping: A review. J. Plant Ecol. 2008, 1, 9–23. [Google Scholar] [CrossRef]
- Griffiths, P.; Kuemmerle, T.; Baumann, M.; Radeloff, V.C.; Abrudan, I.V.; Lieskovsky, J.; Munteanu, C.; Ostapowicz, K.; Hostert, P. Forest disturbances, forest recovery, and changes in forest types across the carpathian ecoregion from 1985 to 2010 based on landsat image composites. Remote Sens. Environ. 2014, 151, 72–88. [Google Scholar] [CrossRef]
- Pimple, U.; Sitthi, A.; Simonetti, D.; Pungkul, S.; Leadprathom, K.; Chidthaisong, A. Topographic correction of Landsat TM-5 and Landsat OLI-8 imagery to improve the performance of forest classification in the mountainous terrain of Northeast Thailand. Sustainability 2017, 9, 258. [Google Scholar] [CrossRef]
- Yin, H.; Khamzina, A.; Pflugmacher, D.; Martius, C. Forest cover mapping in post-Soviet Central Asia using multi-resolution remote sensing imagery. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Immitzer, M.; Böck, S.; Einzmann, K.; Vuolo, F.; Pinnel, N.; Wallner, A.; Atzberger, C. Fractional cover mapping of spruce and pine at 1 ha resolution combining very high and medium spatial resolution satellite imagery. Remote Sens. Environ. 2017, 204, 690–703. [Google Scholar] [CrossRef]
- Schlerf, M.; Atzberger, C.; Hill, J. Tree species and age class mapping in a Central European woodland using optical remote sensing imagery and orthophoto derived stem density—Performance of multispectral and hyperspectral sensors. In Proceedings of the 22nd EARSeL Symposium Geoinformation for European-Wide Integration, Prague, Czech Republic, 4–6 June 2002; pp. 413–418. [Google Scholar]
- Townshend, J.R.; Masek, J.G.; Huang, C.; Vermote, E.F.; Gao, F.; Channan, S.; Sexton, J.O.; Feng, M.; Narasimhan, R.; Kim, D.; et al. Global characterization and monitoring of forest cover using Landsat data: Opportunities and challenges. Int. J. Digit. Earth 2012, 5, 373–397. [Google Scholar] [CrossRef]
- Dudley, K.L.; Dennison, P.E.; Roth, K.L.; Roberts, D.A.; Coates, A.R. A multi-temporal spectral library approach for mapping vegetation species across spatial and temporal phenological gradients. Remote Sens. Environ. 2015, 167, 121–134. [Google Scholar] [CrossRef]
- George, R.; Padalia, H.; Kushwaha, S.P.S. Forest tree species discrimination in western Himalaya using EO-1 Hyperion. Int. J. Appl. Earth Obs. Geoinf. 2014, 28, 140–149. [Google Scholar] [CrossRef]
- Shukla, A.; Kot, R. An Overview of Hyperspectral Remote Sensing and its applications in various Disciplines. Int. J. Appl. Sci. 2016, 05, 85–90. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Holmgren, J. Identifying species of individual trees using airborne laser scanner. Remote Sens. Environ. 2004, 90, 415–423. [Google Scholar] [CrossRef]
- Holmgren, J.; Persson, A.; Söderman, U. Species Identification of Individual Trees by Combining High Resolution LiDAR Data with Multi-Spectral Images. Int. J. Remote Sens. 2008, 29, 1537–1552. [Google Scholar] [CrossRef]
- Åkerblom, M.; Raumonen, P.; Mäkipää, R.; Kaasalainen, M. Remote Sensing of Environment. Remote Sens. Environ. 2017, 191, 1–12. [Google Scholar] [CrossRef]
- Krahwinkler, P.; Rossmann, J. Tree species classification and input data evaluation. Eur. J. Remote Sens. 2013, 46, 535–549. [Google Scholar] [CrossRef]
- Gudex-Cross, D.; Pontius, J.; Adams, A. Enhanced forest cover mapping using spectral unmixing and object-based classification of multi-temporal Landsat imagery. Remote Sens. Environ. 2017, 196, 193–204. [Google Scholar] [CrossRef]
- Meng, J.; Li, S.; Wang, W.; Liu, Q.; Xie, S.; Ma, W.; Ganguly, S.; Tucker, C.; Roy, S.; Thenkabail, P.S. Mapping Forest Health Using Spectral and Textural Information Extracted from SPOT-5 Satellite Images. Remote Sens. 2016, 8, 719. [Google Scholar] [CrossRef]
- Clark, M.L.; Roberts, D.A.; Clark, D.B. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Waser, L.T.; Küchler, M.; Jütte, K.; Stampfer, T. Evaluating the potential of worldview-2 data to classify tree species and different levels of ash mortality. Remote Sens. 2014, 6, 4515–4545. [Google Scholar] [CrossRef]
- Key, T.; Warner, T.A.; McGraw, J.B.; Fajvan, M.A. A comparison of multispectral and multitemporal information in high spatial resolution imagery for classification of individual tree species in a temperate hardwood forest. Remote Sens. Environ. 2001, 75, 100–112. [Google Scholar] [CrossRef]
- Delpierre, N.; Vitasse, Y.; Chuine, I.; Guillemot, J.; Bazot, S.; Rutishauser, T.; Rathgeber, C.B.K. Temperate and boreal forest tree phenology: from organ-scale processes to terrestrial ecosystem models. Ann. For. Sci. 2016, 73, 5–25. [Google Scholar] [CrossRef] [Green Version]
- Klosterman, S.; Richardson, A.D. Observing spring and fall phenology in a deciduous forest with aerial drone imagery. Sensors 2017, 17, 2852. [Google Scholar] [CrossRef]
- Vilhar, U.Š.; Beuker, E.; Mizunuma, T.; Skudnik, M.; Lebourgeois, F.; Soudani, K.; Wilkinson, M. Tree Phenology. Dev. Environ. Sci. 2013, 12, 169–182. [Google Scholar] [CrossRef]
- Junker, L.V.; Ensminger, I. Relationship between leaf optical properties, chlorophyll fluorescence and pigment changes in senescing Acer saccharum leaves. Tree Physiol. 2016, 36, 694–711. [Google Scholar] [CrossRef] [Green Version]
- Schieber, B.; Janík, R.; Snopková, Z. Phenology of four broad-leaved forest trees in a submountain beech forest. J. For. Sci. 2009, 55, 15–22. [Google Scholar] [CrossRef] [Green Version]
- Hill, R.A.; Wilson, A.K.; George, M.; Hinsley, S.A. Mapping tree species in temperate deciduous woodland using time-series multi-spectral data. Appl. Veg. Sci. 2010, 13, 86–99. [Google Scholar] [CrossRef]
- Bayr, C.; Gallaun, H.; Kleb, U.; Kornberger, B.; Steinegger, M.; Winter, M. Satellite-based forest monitoring: Spatial and temporal forecast of growing index and short-wave infrared band. Geospat. Health 2016, 11, 31–42. [Google Scholar] [CrossRef]
- Addabbo, P.; Focareta, M.; Marcuccio, S.; Votto, C.; Ullo, S. Contribution of Sentinel-2 data for applications in vegetation monitoring. Acta Imeko 2016, 5, 44–54. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W.; Gitelson, A.A. Remote estimation of crop and grass chlorophyll and nitrogen content using red-edge bands on Sentinel-2 and -3. Int. J. Appl. Earth Obs. Geoinf. 2013, 23, 334–343. [Google Scholar] [CrossRef]
- Immitzer, M.; Vuolo, F.; Atzberger, C. First Experience with Sentinel-2 Data for Crop and Tree Species Classifications in Central Europe. Remote Sens. 2016, 8, 166. [Google Scholar] [CrossRef]
- Karasiak, N.; Sheeren, D.; Fauvel, M.; Willm, J.; Dejoux, J.-F.; Monteil, C. Mapping Tree Species of Forests in Southwest France using Sentinel-2 Image Time Series. In Proceedings of the 2017 9th International Workshop on the Analysis of Multitemporal Remote Sensing Images (MultiTemp), Brugge, Belgium, 27–29 June 2017; pp. 1–4. [Google Scholar]
- Persson, M.; Lindberg, E.; Reese, H. Tree Species Classification with Multi-Temporal Sentinel-2 Data. Remote Sens. 2018, 10, 1794. [Google Scholar] [CrossRef]
- Wessel, M.; Brandmeier, M.; Tiede, D. Evaluation of different machine learning algorithms for scalable classification of tree types and tree species based on Sentinel-2 data. Remote Sens. 2018, 10. [Google Scholar] [CrossRef]
- Godzik, B.; Grodzińska, K. Vegetation of the selected forest stands in the polish carpathian mountains—Changing in time. Ekol. Bratislava 2008, 27, 300–315. [Google Scholar]
- Marszałek, E. Gospodarka leśna w karpackiej części Regionalnej Dyrekcji Lasów Państwowych w Krośnie i jej wpływ na ochronę przyrody [Forest management in the Carpathian part of the Regional Directorate of State Forests in Krosno and its influence on nature protection]. Rocz. Bieszczadzkie 2011, 19, 59–75. [Google Scholar]
- Nadleśnictwo Baligród. Available online: http://www.baligrod.krosno.lasy.gov.pl (accessed on 30 March 2019).
- Bank Danych o Lasach. Available online: https://www.bdl.lasy.gov.pl/portal/ (accessed on 30 March 2019).
- Calle, M.L.; Urrea, V. Letter to the editor: Stability of Random Forest importance measures. Brief. Bioinform. 2011, 12, 86–89. [Google Scholar] [CrossRef]
- Mellor, A.; Haywood, A.; Stone, C.; Jones, S. The performance of random forests in an operational setting for large area sclerophyll forest classification. Remote Sens. 2013, 5, 2838–2856. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăgu, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Georganos, S.; Grippa, T.; Vanhuysse, S.; Lennert, M.; Shimoni, M.; Kalogirou, S.; Wolff, E. Less is more: optimizing classification performance through feature selection in a very-high-resolution remote sensing object-based urban application. GISci. Remote Sens. 2018, 55, 221–242. [Google Scholar] [CrossRef]
- Boonprong, S.; Cao, C.; Chen, W.; Bao, S. Random Forest Variable Importance Spectral Indices Scheme for Burnt Forest Recovery Monitoring—Multilevel RF-VIMP. Remote Sens. 2017, 10, 807. [Google Scholar] [CrossRef]
- Behnamian, A.; Millard, K.; Banks, S.N.; White, L.; Richardson, M.; Pasher, J. A Systematic Approach for Variable Selection with Random Forests: Achieving Stable Variable Importance Values. IEEE Geosci. Remote Sens. Lett. 2017, 14, 1988–1992. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Rodriguez-Galiano, V.F.; Ghimire, B.; Rogan, J.; Chica-Olmo, M.; Rigol-Sanchez, J.P. An assessment of the effectiveness of a random forest classifier for land-cover classification. ISPRS J. Photogramm. Remote Sens. 2012, 67, 93–104. [Google Scholar] [CrossRef]
- Gislason, P.O.; Benediktsson, J.A.; Sveinsson, J.R. Random forests for land cover classification. Pattern Recognit. Lett. 2006, 27, 294–300. [Google Scholar] [CrossRef]
- Leutner, B.; Horning, N.; Schwalb-Willmann, J. RStoolbox: Tools for Remote Sensing Data Analysis, R Package Version 0.2.4; 2019. Available online: https://CRAN.R-project.org/package=RStoolbox (accessed on 30 March 2019).
- Olofsson, P.; Foody, G.M.; Herold, M.; Stehman, S.V.; Woodcock, C.E.; Wulder, M.A. Good practices for estimating area and assessing accuracy of land change. Remote Sens. Environ. 2014, 148, 42–57. [Google Scholar] [CrossRef] [Green Version]
- Foody, G.M. Status of land cover classification accuracy assessment. Remote Sens. Environ. 2002, 80, 185–201. [Google Scholar] [CrossRef]
- Louarn, M.L.; Clergeau, P.; Briche, E.; Deschamps-Cottin, M. “Kill two birds with one stone”: Urban tree species classification using Bi-Temporal pléiades images to study nesting preferences of an invasive bird. Remote Sens. 2017, 9. [Google Scholar] [CrossRef]
- Wolter, P.T.; Mladenoff, D.J.; Host, G.E.; Crow, T.R. Improved Forest Classification in the Northern Lake States Using Multi-Temporal Landsat Imagery. Photogramm. Eng. Remote Sens. 1995, 61, 1129–1143. [Google Scholar]
- Pasquarella, V.J.; Holden, C.E.; Woodcock, C.E. Improved mapping of forest type using spectral-temporal Landsat features. Remote Sens. Environ. 2018, 210, 193–207. [Google Scholar] [CrossRef]
- Schriever, J.R.; Congalton, R.G. Evaluating Seasonal Variability as an Aid to Cover-Type Mapping from Landsat Thematic Mapper Data in the Northeast. Photogramm. Eng. Remote Sens. 1995, 61, 321–327. [Google Scholar]
- Lisein, J.; Michez, A.; Claessens, H.; Lejeune, P. Discrimination of deciduous tree species from time series of unmanned aerial system imagery. PLoS ONE 2015, 10, 1–20. [Google Scholar] [CrossRef]
- Hovi, A.; Raitio, P.; Rautiainen, M. A spectral analysis of 25 boreal tree species. Silva Fenn. 2017, 51, 1–16. [Google Scholar] [CrossRef]
- Gamon, J.A.; Huemmrich, K.F.; Wong, C.Y.S.; Ensminger, I.; Garrity, S.; Hollinger, D.Y.; Noormets, A.; Peñuelas, J. A remotely sensed pigment index reveals photosynthetic phenology in evergreen conifers. Proc. Natl. Acad. Sci. USA 2016, 113, 13087–13092. [Google Scholar] [CrossRef] [Green Version]
- Niemann, O. Remote sensing of forest stand age using airborne spectrometer data. Photogramm. Eng. Remote Sens. 1995, 61, 1119–1126. [Google Scholar]
- Puletti, N.; Chianucci, F.; Castaldi, C. Use of Sentinel-2 for forest classification in Mediterranean environments. Ann. Silvic. Res. 2017, 0, 1–7. [Google Scholar] [CrossRef]
- Dymond, C.C.; Mladenoff, D.J.; Radeloff, V.C. Phenological differences in Tasseled Cap indices improve deciduous forest classification. Remote Sens. Environ. 2002, 80, 460–472. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree Species Classification with Random Forest Using Very High Spatial Resolution 8-Band WorldView-2 Satellite Data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef] [Green Version]
- Carleer, A.; Wolff, E. Exploitation of Very High Resolution Satellite Data for Tree Species Identification. Photogramm. Eng. Remote Sens. 2004, 70, 135–140. [Google Scholar] [CrossRef]
- Bolyn, C.; Michez, A.; Gaucher, P.; Lejeune, P.; Bonnet, S. Forest mapping and species composition using supervised per pixel classification of Sentinel-2 imagery. Biotechnol. Agron. Soc. Environ. 2018, 22. [Google Scholar] [CrossRef]
- Ferreira, M.P.; Zortea, M.; Zanotta, D.C.; Féret, J.B.; Shimabukuro, Y.E.; Filho, C.R.S. On the use of shortwave infrared for tree species discrimination in tropical semideciduous forest. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. ISPRS Arch. 2015, 40, 473–476. [Google Scholar] [CrossRef]
- Adamczyk, J.; Osberger, A. Red-edge vegetation indices for detecting and assessing disturbances in Norway spruce dominated mountain forests. Int. J. Appl. Earth Obs. Geoinf. 2015, 37, 90–99. [Google Scholar] [CrossRef]
- Forkuor, G.; Dimobe, K.; Serme, I.; Tondoh, J.E. Landsat-8 vs. Sentinel-2: examining the added value of sentinel-2’s red-edge bands to land-use and land-cover mapping in Burkina Faso. GISci. Remote Sens. 2018, 55, 331–354. [Google Scholar] [CrossRef]
- Sommer, C.; Holzwarth, S.; Heiden, U.; Heurich, M. Feature-Based Tree Species Classification Using Hyperspectral and Lidar Data. EARSeL eProc. Spec. Issue 2015, 14, 49–70. [Google Scholar] [CrossRef]
- Sun, Y.; Wong, A.K.C.; Kamel, M.S. Classification of imbalanced data: A review. Int. J. Pattern Recognit. Artif. Intell. 2009, 23, 687–719. [Google Scholar] [CrossRef]
- Abdollahnejad, A.; Panagiotidis, D.; Joybari, S.S.; Surovỳ, P. Prediction of dominant forest tree species using quickbird and environmental data. Forests 2017, 8, 42. [Google Scholar] [CrossRef]
- Isaacson, B.N.; Serbin, S.P.; Townsend, P.A. Detection of relative differences in phenology of forest species using Landsat and MODIS. Landsc. Ecol. 2012, 27, 529–543. [Google Scholar] [CrossRef]
- Leckie, D.G.; Tinis, S.; Nelson, T.; Burnett, C.; Gougeon, F.A.; Cloney, E.; Paradine, D. Issues in species classification of trees in old growth conifer stands. Can. J. Remote Sens. 2005, 31, 175–190. [Google Scholar] [CrossRef] [Green Version]
- Stoffels, J.; Mader, S.; Hill, J.; Werner, W.; Ontrup, G. Satellite-based stand-wise forest cover type mapping using a spatially adaptive classification approach. Eur. J. For. Res. 2012, 131, 1071–1089. [Google Scholar] [CrossRef]









| Tree Species | Number of Polygons | Area [ha] |
|---|---|---|
| Common beech | 76 | 578.7 |
| Silver birch | 6 | 4.9 |
| Common hornbeam | 6 | 14.65 |
| Silver fir | 59 | 127.9 |
| Sycamore maple | 9 | 18.9 |
| European larch | 8 | 12.5 |
| Grey alder | 8 | 9.1 |
| Scots pine | 37 | 35.4 |
| Norway spruce | 11 | 12.6 |
| Total | 220 | 814.6 |
| Number of Images | Combination |
|---|---|
| Two | 05-May/14-Oct |
| 30-Apr/17-Oct | |
| 14-Oct/17-Oct | |
| Three | 05-May/06-Jun/14-Oct |
| 30-Apr/14-Oct/17-Oct | |
| 05-May/14-Oct/17-Oct | |
| 05-Apr/05-May/08-Nov | |
| Four | 05-Apr/05-May/14-Oct/08-Nov |
| 30-Apr/05-May/14-Oct/17-Oct | |
| 30-Apr/05-May/17-Oct/08-Nov | |
| Five | 30-Apr/05-May/14-Oct/17-Oct/08-Nov |
| Eighteen | All images |
| Reference | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | Total | ||
| Map | Beech (1) | 932 | 0 | 4 | 1 | 18 | 0 | 0 | 3 | 0 | 958 |
| Birch (2) | 0 | 5 | 9 | 0 | 0 | 0 | 5 | 6 | 0 | 25 | |
| Hornbeam (3) | 4 | 6 | 120 | 0 | 0 | 0 | 0 | 1 | 0 | 131 | |
| Fir (4) | 0 | 0 | 2 | 830 | 0 | 52 | 15 | 0 | 21 | 920 | |
| Sycamore (5) | 4 | 1 | 4 | 0 | 47 | 0 | 1 | 6 | 0 | 63 | |
| Spruce (6) | 0 | 0 | 0 | 0 | 0 | 64 | 0 | 0 | 0 | 64 | |
| Larch (7) | 4 | 6 | 1 | 0 | 0 | 0 | 62 | 1 | 0 | 74 | |
| Grey alder (8) | 4 | 0 | 0 | 0 | 3 | 0 | 0 | 9 | 0 | 16 | |
| Pine (9) | 3 | 0 | 0 | 1 | 0 | 4 | 0 | 0 | 174 | 182 | |
| Total | 951 | 18 | 140 | 832 | 68 | 120 | 83 | 26 | 195 | 2433 | |
| Prod. Acc. | 98.0 | 27.8 | 85.7 | 99.8 | 69.1 | 53.3 | 74.7 | 34.6 | 89.2 | ||
| User Acc. | 97.3 | 20.0 | 91.6 | 90.2 | 74.6 | 100 | 83.8 | 56.3 | 95.6 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grabska, E.; Hostert, P.; Pflugmacher, D.; Ostapowicz, K. Forest Stand Species Mapping Using the Sentinel-2 Time Series. Remote Sens. 2019, 11, 1197. https://doi.org/10.3390/rs11101197
Grabska E, Hostert P, Pflugmacher D, Ostapowicz K. Forest Stand Species Mapping Using the Sentinel-2 Time Series. Remote Sensing. 2019; 11(10):1197. https://doi.org/10.3390/rs11101197
Chicago/Turabian StyleGrabska, Ewa, Patrick Hostert, Dirk Pflugmacher, and Katarzyna Ostapowicz. 2019. "Forest Stand Species Mapping Using the Sentinel-2 Time Series" Remote Sensing 11, no. 10: 1197. https://doi.org/10.3390/rs11101197
APA StyleGrabska, E., Hostert, P., Pflugmacher, D., & Ostapowicz, K. (2019). Forest Stand Species Mapping Using the Sentinel-2 Time Series. Remote Sensing, 11(10), 1197. https://doi.org/10.3390/rs11101197