Classification of Mangrove Species Using Combined WordView-3 and LiDAR Data in Mai Po Nature Reserve, Hong Kong
Abstract
:1. Introduction
2. Materials and Methods
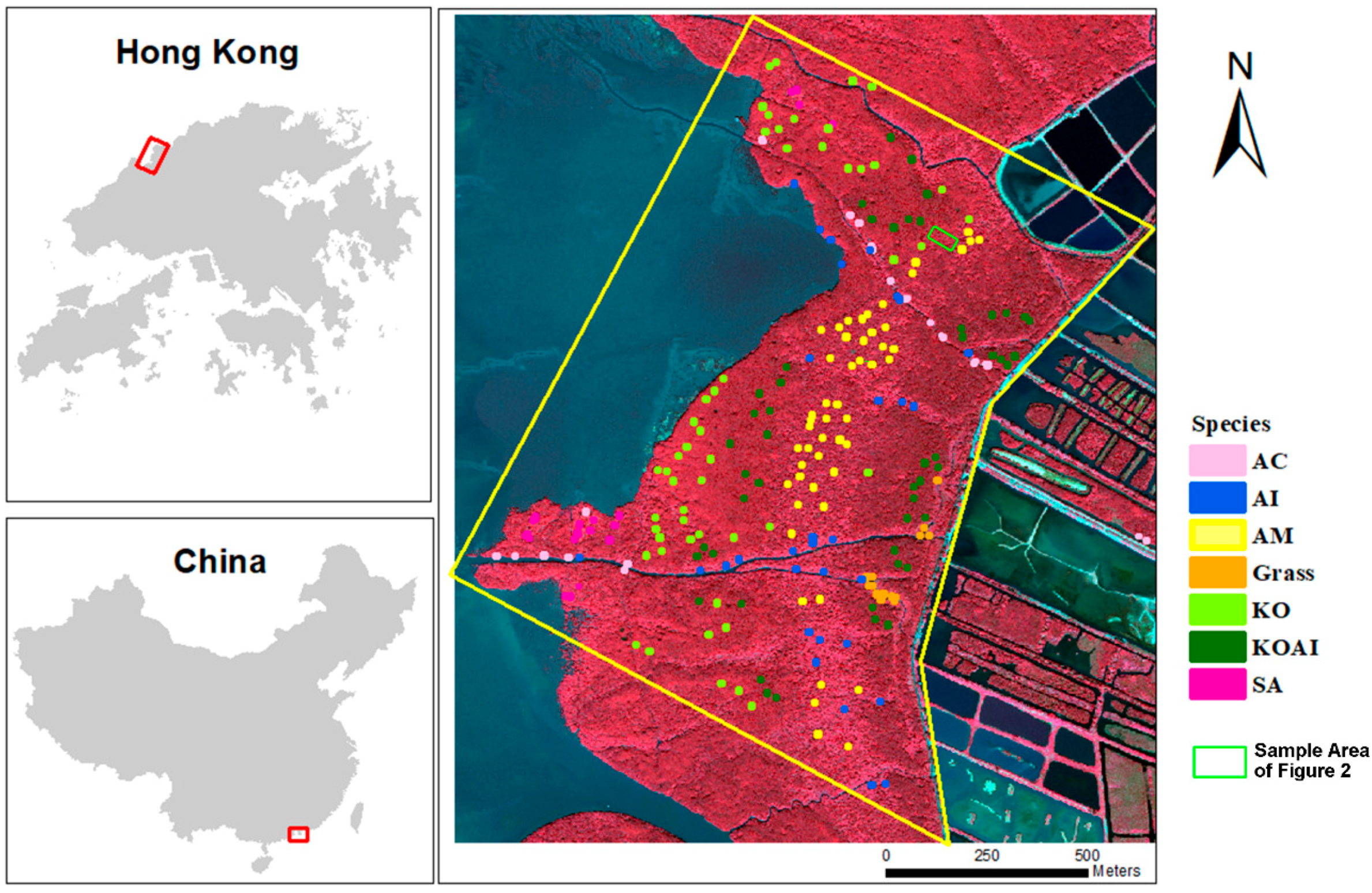
2.1. Study Area
2.2. Remotely Sensed Data and Preprocessing
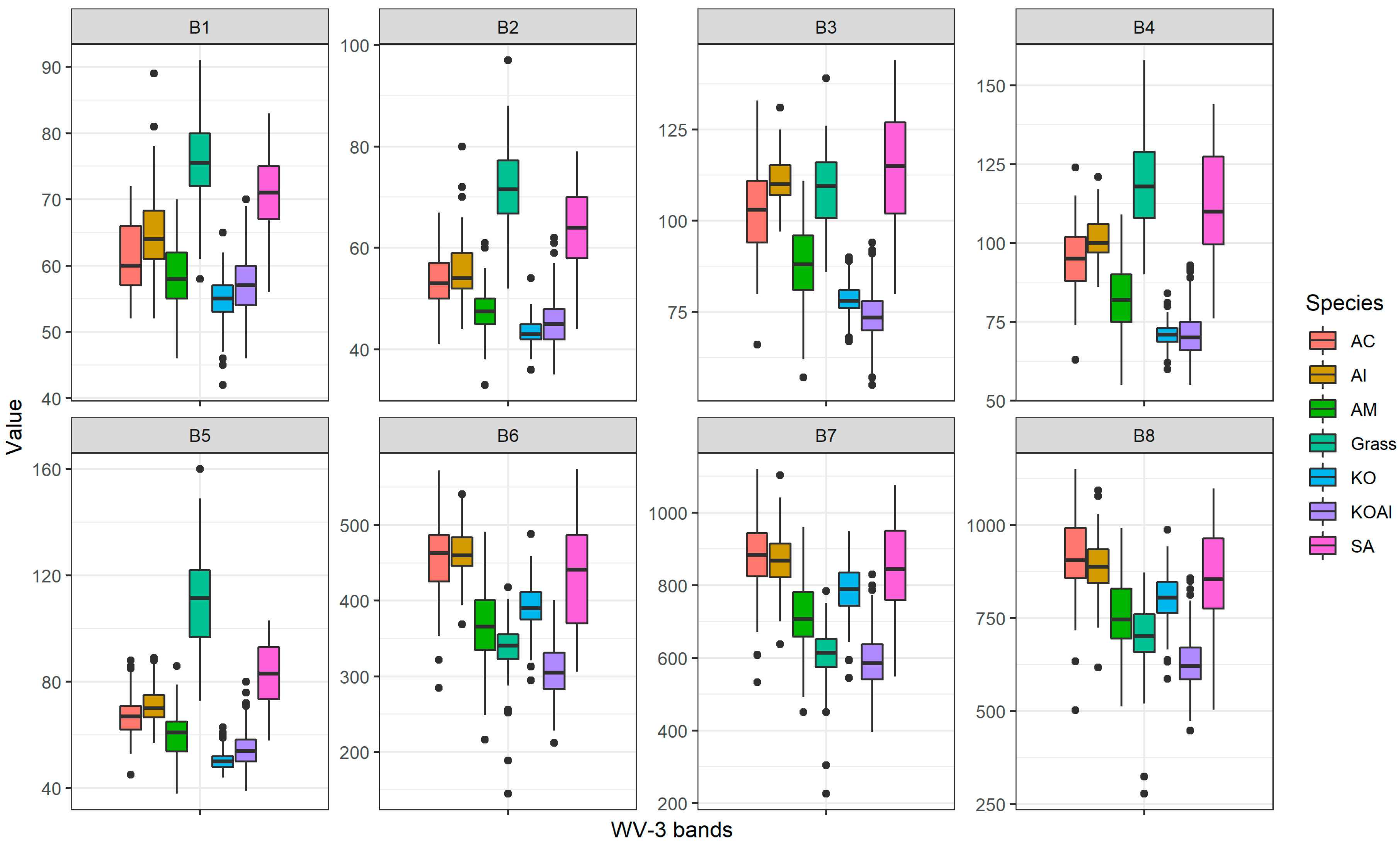
2.2.1. WorldView-3 Image
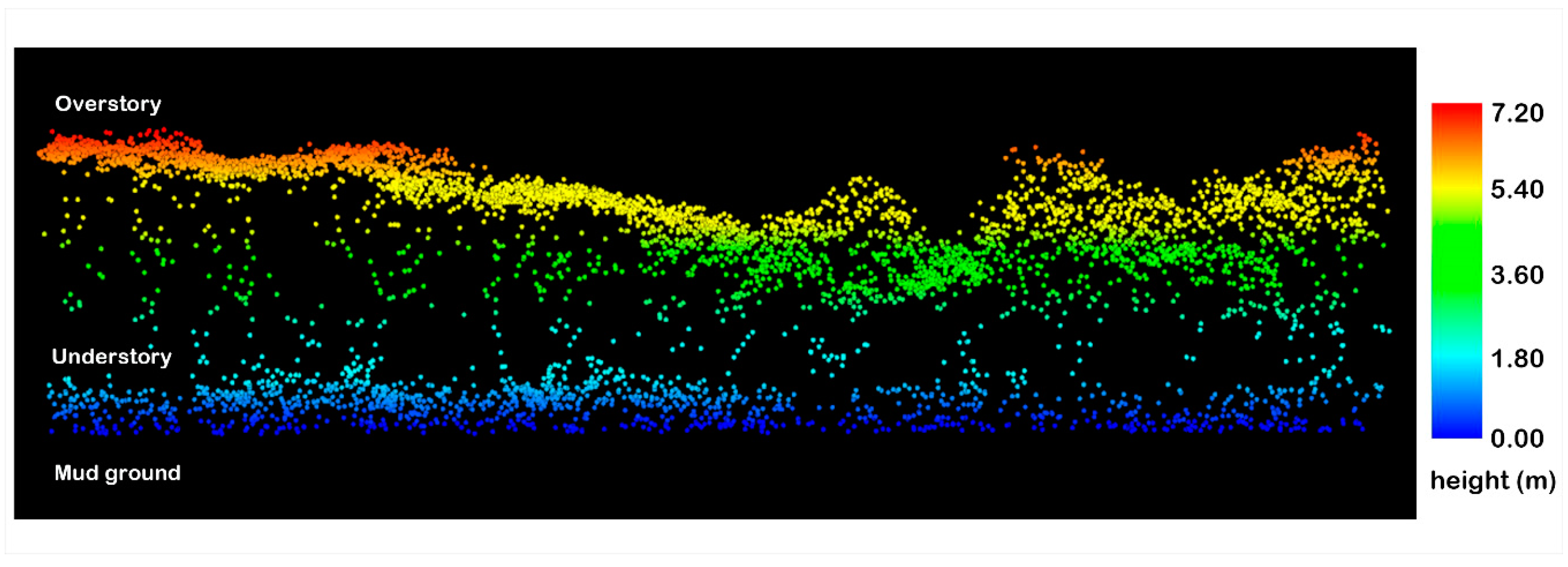
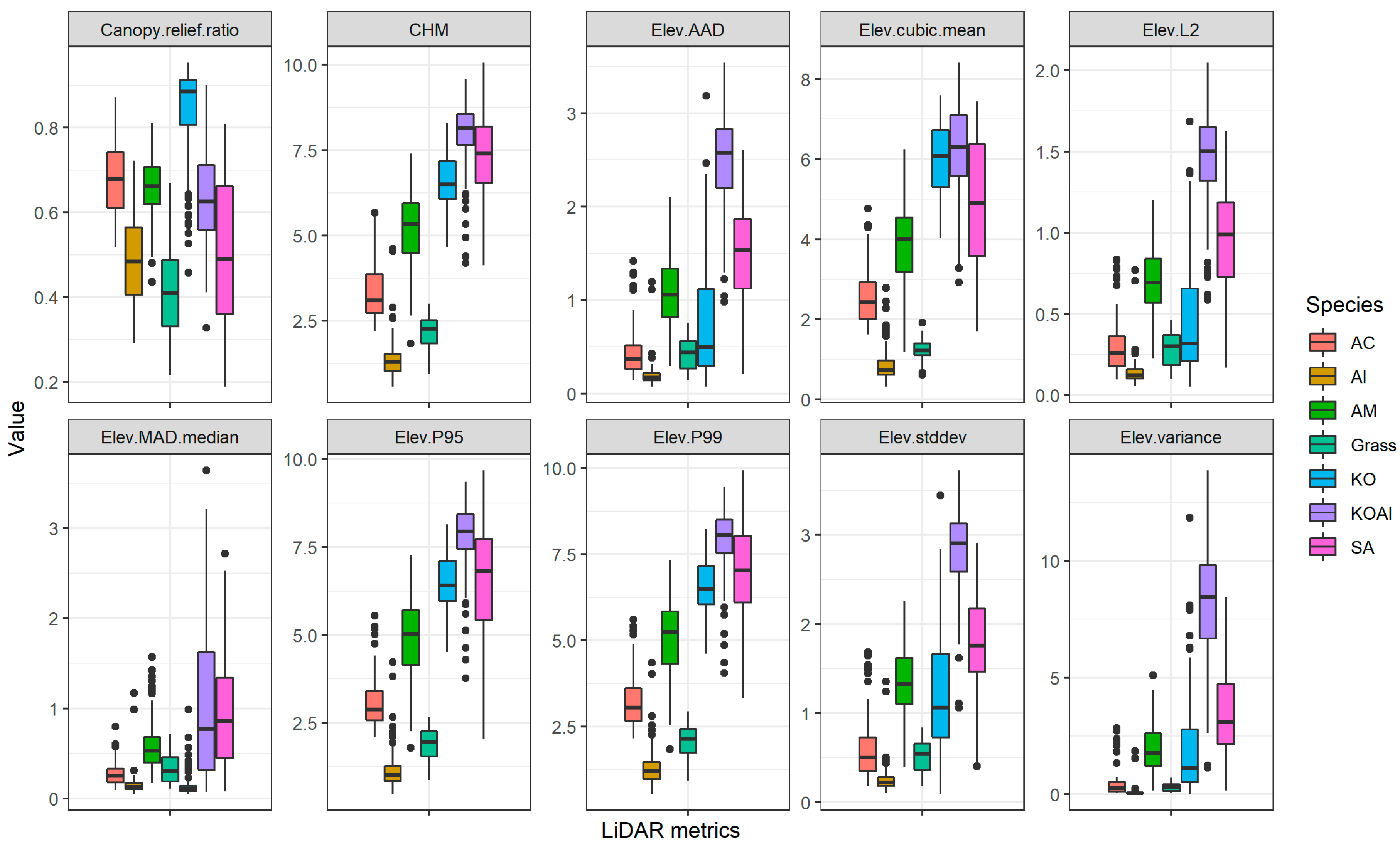
2.2.2. Airborne LiDAR Data
2.3. Field Survey and Reference Data Collection
2.4. Feature Generation and Selection
2.5. Classification and Validation
2.5.1. Support Vector Machine Classifier
2.5.2. Random Forest Classifier
2.5.3. Validation
3. Results
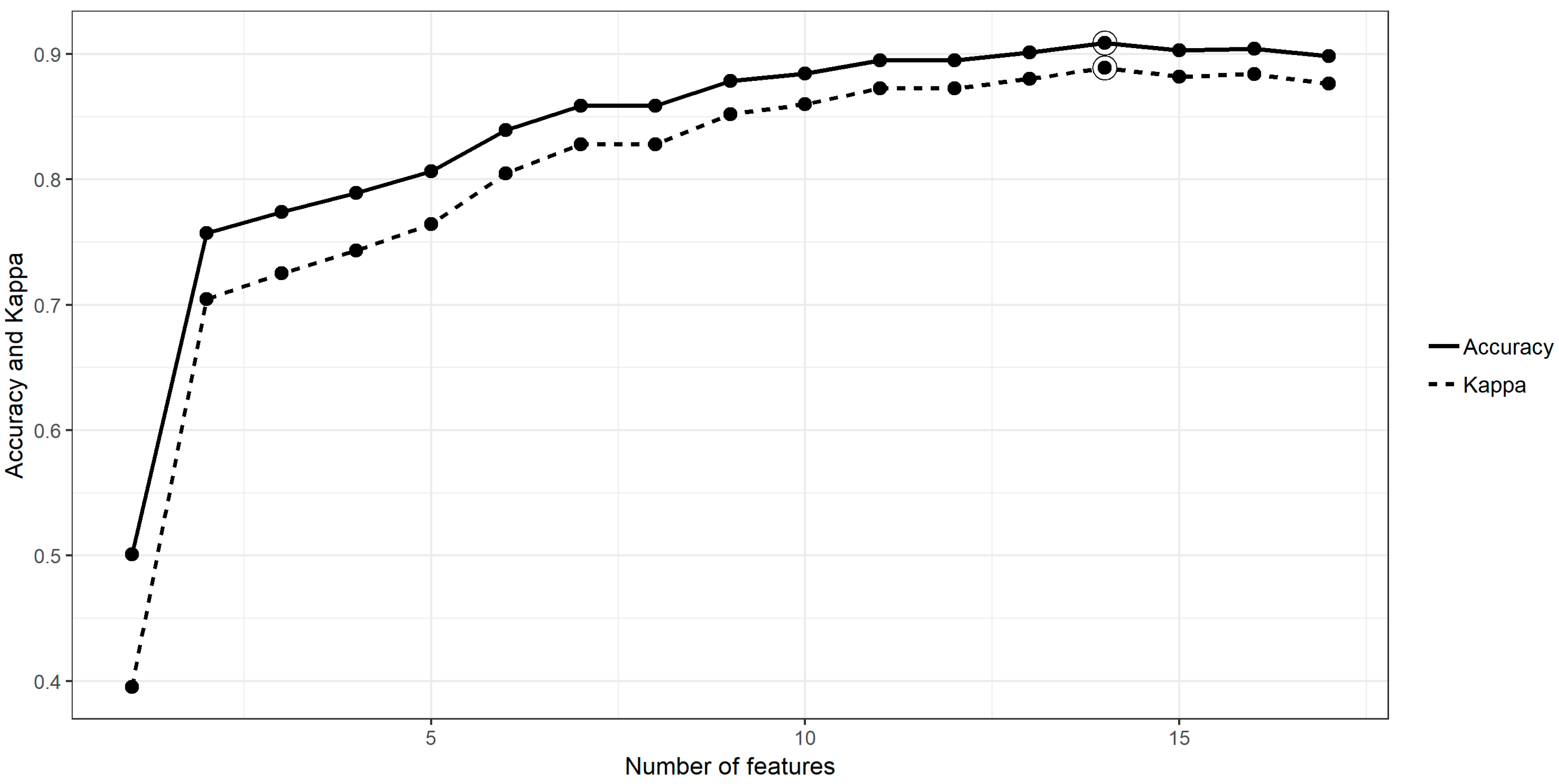
3.1. Feature Selection
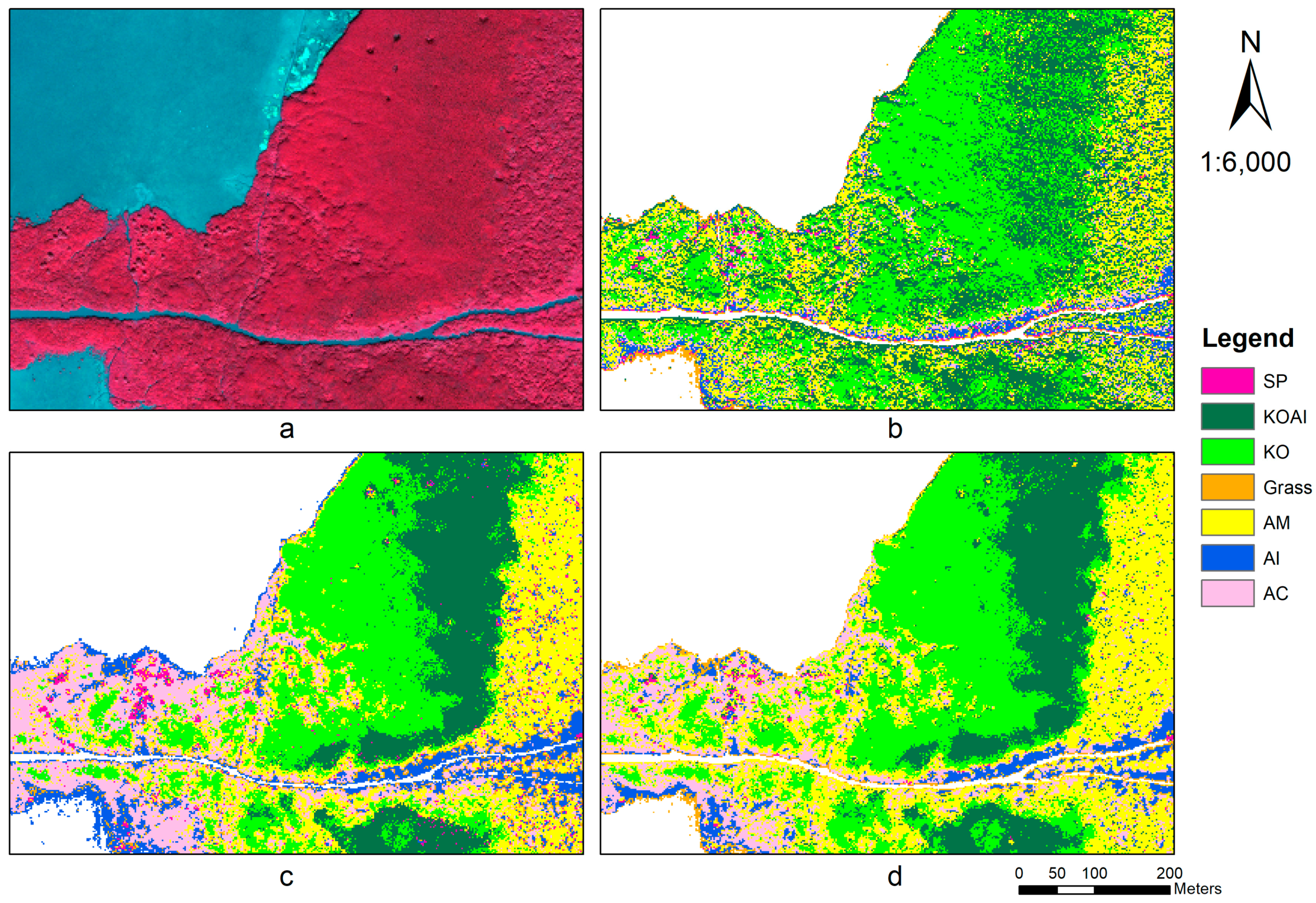
3.2. Classification and Validation
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Abbreviation of LiDAR Metric | Explanation |
|---|---|
| Elev minimum | Elevation minimum |
| CHM | Canopy height model |
| Elev mean | Elevation mean |
| Elev mode | Elevation mode |
| Elev stddev | Elevation standard deviation |
| Elev variance | Elevation variance |
| Elev CV | Elevation coefficient of variation |
| Elev IQ | Elevation 75th percentile minus 25th percentile |
| Elev skewness | Elevation skewness |
| Elev kurtosis | Elevation kurtosis |
| Elev AAD | Elevation average absolute deviation from mean |
| Elev L1, Elev L2,Elev L3,Elev L4 | Elevation L-moments (L1, L2, L3, L4) |
| Elev L CV | Elevation L-moments coefficient of variation |
| Elev L skewness | Elevation L-moments skewness |
| Elev L kurtosis | Elevation L-moments kurtosis |
| Elev P01, Elev P05, Elev 10, Elev P20, Elev P25, Elev P30, Elev P40, Elev P50, Elev P60, Elev P70, Elev P75, Elev P80, Elev 90, Elev P95, Elev P99 | 1st, 5th, 10th 20th, 25th, 30th, 40th, 50th, 60th, 70th, 75th, 80th, 90th, 95th, 99th percentile of elevation |
| Return 1 count | Count of return 1 points |
| Return 2 count | Count of return 2 points |
| canopy cover estimate | Percentage first returns above 1.8 m |
| Percentage all returns above 1.8 | Percentage all returns above 1.8 m |
| (All returns above 1.8)/(Total first returns) × 100 | (All returns above 1.8)/(Total first returns) × 100 |
| First returns above 1.8 | First returns above 1.8 m |
| All returns above 1.8 | All returns above 1.8 m |
| Percentage first returns above mean | Percentage first returns above mean |
| Percentage first returns above mode | Percentage first returns above mode |
| Percentage all returns above mean | Percentage all returns above mean |
| Percentage all returns above mode | Percentage all returns above mode |
| Number of returns above the mean height Number of total first returns × 100 | (All returns above mean)/(Total first returns) × 100 |
| Number of returns above the mode height/Number of total first returns × 100 | (All returns above mode)/(Total first returns) × 100 |
| First returns above mean | First returns above mean |
| First returns above mode | First returns above mode |
| All returns above mean | All returns above mean |
| All returns above mode | All returns above mode |
| Total first returns | Total first returns |
| Total all returns | Total all returns |
| Elev MAD median | Elevation median absolute deviation from the median |
| Elev MAD mode | Elevation median absolute deviation from the mode |
| Canopy relief ratio | Elevation (mean-min)/(max–min) |
| Elev quadratic mean | Elevation quadratic mean |
| Elev cubic mean | Elevation cubic mean |
References
- Tam, N.F.Y.; Wong, Y.-S. Hong Kong Mangroves; City University of Hong Kong Press: Hong Kong, China, 2000; ISBN 9629370557. Available online: https://julac.hosted.exlibrisgroup.com/primo-explore/fulldisplay?docid=CUHK_IZ21810919350003407&context=L&vid=CUHK&lang=en_US&search_scope=Books&adaptor=Local Search Engine&tab=default_tab&query=any,contains, HONG KONG Mangroves&sortby=rank (accessed on 11 September 2019).
- Heenkenda, M.; Joyce, K.; Maier, S.; Bartolo, R. Mangrove Species Identification: Comparing WorldView-2 with Aerial Photographs. Remote Sens. 2014, 6, 6064–6088. [Google Scholar] [CrossRef] [Green Version]
- Heumann, B.W. Satellite remote sensing of mangrove forests: Recent advances and future opportunities. Prog. Phys. Geogr. 2011, 35, 87–108. [Google Scholar] [CrossRef]
- Neukermans, G.; Dahdouh-Guebas, F.; Kairo, J.G.; Koedam, N. Mangrove species and stand mapping in Gazi bay (Kenya) using quickbird satellite imagery. J. Spat. Sci. 2008, 53, 75–86. [Google Scholar] [CrossRef]
- Wang, L.; Sousa, W.P.; Gong, P.; Biging, G.S. Comparison of IKONOS and QuickBird images for mapping mangrove species on the Caribbean coast of Panama. Remote Sens. Environ. 2004, 91, 432–440. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.; Lin, H.; Fang, C. Textural–Spectral Feature-Based Species Classification of Mangroves in Mai Po Nature Reserve from Worldview-3 Imagery. Remote Sens. 2015, 8, 24. [Google Scholar] [CrossRef]
- Ghosh, A.; Joshi, P.K. A comparison of selected classification algorithms for mapping bamboo patches in lower Gangetic plains using very high resolution WorldView 2 imagery. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 298–311. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree Species Classification with Random Forest Using Very High Spatial Resolution 8-Band WorldView-2 Satellite Data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef] [Green Version]
- Sothe, C.; de Almeida, C.M.; Schimalski, M.B.; Liesenberg, V. Integration of Worldview-2 and Lidar Data to MAP a Subtropical Forest Area: Comparison of Machine Learning Algorithms. In Proceedings of the IGARSS 2018—IEEE International Geoscience and Remote Sensing Symposium, Valencia, Spain, 22–27 July 2018; pp. 6207–6210. [Google Scholar]
- Tian, J.; Wang, L.; Li, X.; Gong, H.; Shi, C.; Zhong, R.; Liu, X. Comparison of UAV and WorldView-2 imagery for mapping leaf area index of mangrove forest. Int. J. Appl. Earth Obs. Geoinf. 2017, 61, 22–31. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, X. WorldView-2 Satellite Imagery and Airborne LiDAR Data for Object-Based Forest Species Classification in a Cool Temperate Rainforest Environment. In Developments in Multidimensional Spatial Data Models; Springer: Berlin/Heidelberg, Germany, 2013; pp. 103–122. Available online: http://link.springer.com/10.1007/978-3-642-36379-5_7 (accessed on 11 September 2019).
- Zhu, Y.; Liu, K.; Liu, L.; Myint, S.; Wang, S.; Liu, H.; He, Z.; Zhu, Y.; Liu, K.; Liu, L.; et al. Exploring the Potential of WorldView-2 Red-Edge Band-Based Vegetation Indices for Estimation of Mangrove Leaf Area Index with Machine Learning Algorithms. Remote Sens. 2017, 9, 1060. [Google Scholar] [CrossRef]
- Luo, S.; Wang, C.; Pan, F.; Xi, X.; Li, G.; Nie, S.; Xia, S. Estimation of wetland vegetation height and leaf area index using airborne laser scanning data. Ecol. Indic. 2015, 48, 550–559. [Google Scholar] [CrossRef]
- Hamraz, H.; Contreras, M.A.; Zhang, J. Vertical stratification of forest canopy for segmentation of understory trees within small-footprint airborne LiDAR point clouds. ISPRS J. Photogramm. Remote Sens. 2017, 130, 385–392. [Google Scholar] [CrossRef] [Green Version]
- Cao, L.; Coops, N.C.; Innes, J.L.; Sheppard, S.R.J.; Fu, L.; Ruan, H.; She, G. Estimation of forest biomass dynamics in subtropical forests using multi-temporal airborne LiDAR data. Remote Sens. Environ. 2016, 178, 158–171. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, T.; Skidmore, A.K.; Heurich, M. Important LiDAR metrics for discriminating forest tree species in Central Europe. ISPRS J. Photogramm. Remote Sens. 2018, 137, 163–174. [Google Scholar] [CrossRef]
- Jones, T.G.; Coops, N.C.; Sharma, T. Assessing the utility of airborne hyperspectral and LiDAR data for species distribution mapping in the coastal Pacific Northwest, Canada. Remote Sens. Environ. 2010, 114, 2841–2852. [Google Scholar] [CrossRef]
- Liu, L.; Coops, N.C.; Aven, N.W.; Pang, Y. Mapping urban tree species using integrated airborne hyperspectral and LiDAR remote sensing data. Remote Sens. Environ. 2017, 200, 170–182. [Google Scholar] [CrossRef]
- Chadwick, J. Integrated LiDAR and IKONOS multispectral imagery for mapping mangrove distribution and physical properties. Int. J. Remote Sens. 2011, 32, 6765–6781. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Neumann, C.; Forster, M.; Buddenbaum, H.; Ghosh, A.; Clasen, A.; Joshi, P.K.; Koch, B. Comparison of Feature Reduction Algorithms for Classifying Tree Species With Hyperspectral Data on Three Central European Test Sites. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2547–2561. [Google Scholar] [CrossRef]
- Pal, M.; Foody, G.M. Feature Selection for Classification of Hyperspectral Data by SVM. IEEE Trans. Geosci. Remote Sens. 2010, 48, 2297–2307. [Google Scholar] [CrossRef] [Green Version]
- Guyon, I.; Elisseeff, A. An Introduction to Variable and Feature Selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Cheng, Q.; Varshney, P.K.; Arora, M.K. Logistic Regression for Feature Selection and Soft Classification of Remote Sensing Data. IEEE Geosci. Remote Sens. Lett. 2006, 3, 491–494. [Google Scholar] [CrossRef]
- Tang, Y.; Jing, L.; Li, H.; Liu, Q.; Yan, Q.; Li, X. Bamboo Classification Using WorldView-2 Imagery of Giant Panda Habitat in a Large Shaded Area in Wolong, Sichuan Province, China. Sensors 2016, 16, 1957. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.J.; Moskal, L.M.; Kim, S.-H. Modeling approaches to estimate effective leaf area index from aerial discrete-return LIDAR. Agric. For. Meteorol. 2009, 149, 1152–1160. [Google Scholar] [CrossRef]
- WWF Hong Kong Mai Po Nature Reserve | WWF Hong Kong. Available online: https://www.wwf.org.hk/en/whatwedo/water_wetlands/mai_po_nature_reserve/ (accessed on 29 May 2019).
- AFCD Agriculture, Fisheries and Conservation Department. Available online: https://www.afcd.gov.hk/english/conservation/con_wet/con_wet_look/con_wet_look_gen/con_wet_look_gen.html (accessed on 29 May 2019).
- Waser, L.; Küchler, M.; Jütte, K.; Stampfer, T.; Waser, L.T.; Küchler, M.; Jütte, K.; Stampfer, T. Evaluating the Potential of WorldView-2 Data to Classify Tree Species and Different Levels of Ash Mortality. Remote Sens. 2014, 6, 4515–4545. [Google Scholar] [CrossRef] [Green Version]
- Axelsson, P. DEM generation from laser scanner data using adaptive TIN models. Int. Arch. Photogram. Remote Sens. 2000, 33, 110–117. [Google Scholar]
- Lagomasino, D.; Fatoyinbo, T.; Lee, S.; Feliciano, E.; Trettin, C.; Simard, M. A Comparison of Mangrove Canopy Height Using Multiple Independent Measurements from Land, Air, and Space. Remote Sens. 2016, 8, 327. [Google Scholar] [CrossRef] [PubMed]
- Ballanti, L.; Blesius, L.; Hines, E.; Kruse, B. Tree Species Classification Using Hyperspectral Imagery: A Comparison of Two Classifiers. Remote Sens. 2016, 8, 445. [Google Scholar] [CrossRef]
- Raczko, E.; Zagajewski, B. Comparison of support vector machine, random forest and neural network classifiers for tree species classification on airborne hyperspectral APEX images. Eur. J. Remote Sens. 2017, 50, 144–154. [Google Scholar] [CrossRef] [Green Version]
- Mountrakis, G.; Im, J.; Ogole, C. Support vector machines in remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Fan, R.-E.; Chen, P.-H.; Lin, C.-J. Working Set Selection Using Second Order Information for Training Support Vector Machines. J. Mach. Learn. Res. 2005, 6, 1889–1918. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Pike, R.J.; Wilson, S.E. Elevation-Relief Ratio, Hypsometric Integral, and Geomorphic Area-Altitude Analysis. Geol. Soc. Am. Bull. 1971, 82, 1079–1084. [Google Scholar] [CrossRef]
- DigitalGlobe. The Benefits of the Eight Spectral Bands of WORLDVIEW-2. Available online: www.digitalglobe.com (accessed on 29 May 2019).
- Peñuelas, J.; Filella, I. Visible and near-infrared reflectance techniques for diagnosing plant physiological status. Trends Plant Sci. 1998, 3, 151–156. [Google Scholar] [CrossRef]
- Parker, G.G.; Russ, M.E. The canopy surface and stand development: Assessing forest canopy structure and complexity with near-surface altimetry. For. Ecol. Manag. 2004, 189, 307–315. [Google Scholar] [CrossRef]
- Jia, M.; Zhang, Y.; Wang, Z.; Song, K.; Ren, C. Mapping the distribution of mangrove species in the Core Zone of Mai Po Marshes Nature Reserve, Hong Kong, using hyperspectral data and high-resolution data. Int. J. Appl. Earth Obs. Geoinf. 2014, 33, 226–231. [Google Scholar] [CrossRef]
- Wong, F.K.K.; Fung, T. Combining EO-1 Hyperion and Envisat ASAR data for mangrove species classification in Mai Po Ramsar Site, Hong Kong. Int. J. Remote Sens. 2014, 35, 7828–7856. [Google Scholar] [CrossRef]
| Specification | |
|---|---|
| Flight altitude | 155–270 m, an average of 180 m |
| Footprint diameter | 0.09 m |
| Return density | 20/m2 |
| Number of returns | 1–5 |
| Beam divergence | 0.5 mrad |
| Wavelength | Near-infrared (905 nm) |
| Scanning mechanism | Rotating mirror |
| Vegetation Types | Training | Testing | ||
|---|---|---|---|---|
| Samples | Pixels | Samples | Pixels | |
| Aegiceras corniculatum (AC) | 20 | 49 | 8 | 21 |
| Acanthus ilicifolius (AI) | 22 | 88 | 9 | 36 |
| Avicennia marina (AM) | 36 | 144 | 15 | 60 |
| Gramineae (Grass) | 10 | 40 | 4 | 16 |
| Kandelia obovata (KO) | 36 | 144 | 15 | 60 |
| Kandelia obovata & Acanthus ilicifolius (KOAI) | 25 | 140 | 15 | 60 |
| Sonneratia apetala (SA) | 14 | 55 | 6 | 20 |
| Total | 173 | 660 | 72 | 275 |
| Data | Resolution | Selected Features (Ordering by importance) | Classification Accuracy | |||
|---|---|---|---|---|---|---|
| RF | SVM | |||||
| OA | Kappa | OA | Kappa | |||
| WV-3 MS | 2 m | B3, B6, B4, B5, B7, B2, B8 | 0.70 | 0.63 | 0.72 | 0.66 |
| WV-3 PS | 0.5 m | B3, B6, B5, B7, B4, B8, B2, B1 | 0.68 | 0.61 | 0.68 | 0.61 |
| LiDAR Metric | 2 m | CHM, Elev stddev, Elev variance, Elev P99, Elev AAD, Canopy relief ratio, Elev cubic mean, Elev L2, Elev P95, Elev MAD median | 0.79 | 0.75 | 0.79 | 0.74 |
| LiDAR Metric | 5 m | Elev variance, Elev stddev, CHM, Elev AAD, Canopy relief ratio, Elev L2, Elev skewness, (All returns above mean)/(Total first returns) * 100 | 0.78 | 0.73 | 0.78 | 0.74 |
| WV-3 + LiDAR | 2m | Elev cubic mean, CHM, Canopy relief ratio, Elev P99, Elev P95, B6, Elev stddev, Elev variance, B3, B5, B4, B2, Elev MAD median, B7 | 0.87 | 0.85 | 0.88 | 0.85 |
| RF Classifier | ||||||||
|---|---|---|---|---|---|---|---|---|
| Reference | ||||||||
| Classified | AC | AI | AM | Grass | KO | KOAI | SA | UA |
| AC | 4 | 5 | 0 | 0 | 0 | 0 | 1 | 0.40 |
| AI | 6 | 23 | 1 | 0 | 0 | 0 | 0 | 0.77 |
| AM | 5 | 7 | 38 | 3 | 3 | 5 | 0 | 0.62 |
| Grass | 0 | 0 | 0 | 13 | 0 | 0 | 1 | 0.93 |
| KO | 6 | 0 | 4 | 0 | 50 | 8 | 0 | 0.74 |
| KOAI | 0 | 1 | 16 | 0 | 7 | 47 | 2 | 0.64 |
| SA | 0 | 0 | 1 | 0 | 0 | 0 | 18 | 0.95 |
| PA | 0.19 | 0.64 | 0.63 | 0.81 | 0.83 | 0.78 | 0.82 | |
| SVM Classifier | ||||||||
| Reference | ||||||||
| Classified | AC | AI | AM | Grass | KO | KOAI | SA | UA |
| AC | 5 | 7 | 0 | 0 | 0 | 0 | 1 | 0.38 |
| AI | 4 | 24 | 1 | 0 | 0 | 0 | 1 | 0.80 |
| AM | 4 | 4 | 37 | 1 | 3 | 7 | 0 | 0.66 |
| Grass | 0 | 0 | 0 | 15 | 0 | 0 | 0 | 1.00 |
| KO | 7 | 0 | 1 | 0 | 51 | 5 | 0 | 0.80 |
| KOAI | 1 | 1 | 20 | 0 | 6 | 48 | 2 | 0.62 |
| SA | 0 | 0 | 1 | 0 | 0 | 0 | 18 | 0.95 |
| PA | 0.24 | 0.67 | 0.62 | 0.94 | 0.85 | 0.8 | 0.82 | |
| RF Classifier | ||||||||
|---|---|---|---|---|---|---|---|---|
| Reference | ||||||||
| Classified | AC | AI | AM | Grass | KO | KOAI | SA | UA |
| AC | 12 | 4 | 3 | 0 | 0 | 0 | 0 | 0.63 |
| AI | 1 | 26 | 0 | 6 | 0 | 0 | 0 | 0.79 |
| AM | 3 | 2 | 53 | 1 | 3 | 3 | 4 | 0.77 |
| Grass | 0 | 4 | 0 | 9 | 0 | 0 | 1 | 0.64 |
| KO | 3 | 0 | 3 | 0 | 52 | 1 | 1 | 0.87 |
| KOAI | 0 | 0 | 0 | 0 | 5 | 53 | 3 | 0.87 |
| SA | 2 | 0 | 1 | 0 | 0 | 3 | 13 | 0.68 |
| PA | 0.57 | 0.72 | 0.88 | 0.56 | 0.87 | 0.88 | 0.59 | |
| SVM Classifier | ||||||||
| Reference | ||||||||
| Classified | AC | AI | AM | Grass | KO | KOAI | SA | UA |
| AC | 9 | 3 | 0 | 1 | 0 | 0 | 1 | 0.64 |
| AI | 1 | 29 | 0 | 6 | 0 | 0 | 0 | 0.81 |
| AM | 6 | 2 | 60 | 2 | 11 | 4 | 4 | 0.67 |
| Grass | 2 | 2 | 0 | 7 | 0 | 0 | 1 | 0.58 |
| KO | 3 | 0 | 0 | 0 | 48 | 1 | 4 | 0.86 |
| KOAI | 0 | 0 | 0 | 0 | 1 | 54 | 2 | 0.95 |
| SA | 0 | 0 | 0 | 0 | 0 | 1 | 10 | 0.91 |
| PA | 0.43 | 0.81 | 1.00 | 0.44 | 0.8 | 0.9 | 0.45 | |
| RF Classifier | ||||||||
|---|---|---|---|---|---|---|---|---|
| Reference | ||||||||
| Classified | AC | AI | AM | Grass | KO | KOAI | SA | UA |
| AC | 14 | 1 | 1 | 0 | 3 | 0 | 2 | 0.74 |
| AI | 3 | 28 | 2 | 3 | 0 | 0 | 0 | 0.93 |
| AM | 1 | 0 | 57 | 0 | 2 | 0 | 0 | 0.84 |
| Grass | 0 | 0 | 1 | 14 | 0 | 0 | 1 | 0.82 |
| KO | 0 | 0 | 3 | 0 | 55 | 2 | 0 | 0.90 |
| KOAI | 0 | 0 | 4 | 0 | 1 | 54 | 1 | 0.93 |
| SA | 1 | 1 | 0 | 0 | 0 | 2 | 18 | 0.82 |
| PA | 0.67 | 0.78 | 0.95 | 0.88 | 0.92 | 0.9 | 0.82 | |
| SVM Classifier | ||||||||
| Reference | ||||||||
| Classified | AC | AI | AM | Grass | KO | KOAI | SA | UA |
| AC | 12 | 3 | 2 | 0 | 1 | 0 | 0 | 0.67 |
| AI | 0 | 30 | 0 | 0 | 0 | 0 | 0 | 1.00 |
| AM | 4 | 3 | 57 | 1 | 1 | 4 | 1 | 0.80 |
| Grass | 0 | 0 | 0 | 14 | 0 | 0 | 0 | 1.00 |
| KO | 4 | 0 | 1 | 0 | 55 | 1 | 0 | 0.90 |
| KOAI | 0 | 0 | 0 | 0 | 3 | 55 | 2 | 0.92 |
| SA | 1 | 0 | 0 | 1 | 0 | 0 | 19 | 0.90 |
| PA | 0.57 | 0.83 | 0.95 | 0.88 | 0.92 | 0.92 | 0.86 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Wong, F.K.K.; Fung, T. Classification of Mangrove Species Using Combined WordView-3 and LiDAR Data in Mai Po Nature Reserve, Hong Kong. Remote Sens. 2019, 11, 2114. https://doi.org/10.3390/rs11182114
Li Q, Wong FKK, Fung T. Classification of Mangrove Species Using Combined WordView-3 and LiDAR Data in Mai Po Nature Reserve, Hong Kong. Remote Sensing. 2019; 11(18):2114. https://doi.org/10.3390/rs11182114
Chicago/Turabian StyleLi, Qiaosi, Frankie Kwan Kit Wong, and Tung Fung. 2019. "Classification of Mangrove Species Using Combined WordView-3 and LiDAR Data in Mai Po Nature Reserve, Hong Kong" Remote Sensing 11, no. 18: 2114. https://doi.org/10.3390/rs11182114





