The Influence of Vegetation Characteristics on Individual Tree Segmentation Methods with Airborne LiDAR Data
Abstract
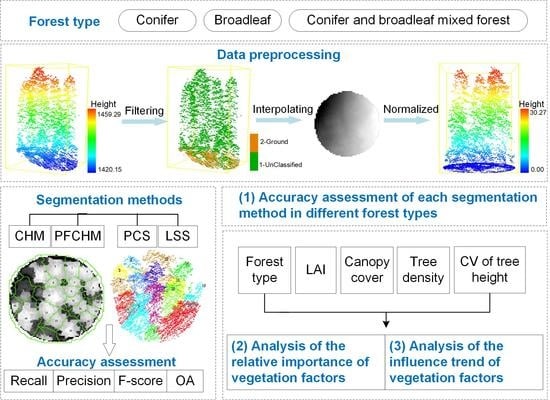
:1. Introduction
2. Data and Methods
2.1. Study Area and Data
2.2. Data Preprocessing
2.3. Segmentation Methods
2.4. Accuracy Assessment of Different Individual Tree Segmentation Methods
2.5. Extraction of Vegetation Characteristics
2.6. The Relative Importance of Vegetation Characteristics
2.7. Trends in the Influence of Vegetation Characteristics
3. Results
3.1. Accuracy Assessment of Different Segmentation Methods in Different Forest Types
3.2. Analysis of the Relative Importance of Vegetation Characteristics
3.3. Analysis of the Influence Trend of Vegetation Characteristics
4. Discussion
4.1. Differences in Segmentation Accuracies of Different Methods in Three Forest Types
4.2. The Influence of Vegetation Characteristics on Segmentation Accuracies
4.3. Limitations of the Present Study
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Craw, D.; Upton, P.; Burridge, C.P.; Wallis, G.P.; Waters, J.M. Rapid biological speciation driven by tectonic evolution in New Zealand. Nat. Geosci. 2016, 9, 140–144. [Google Scholar] [CrossRef]
- Siepielski, A.M.; Morrissey, M.B.; Buoro, M.; Carlson, S.M.; Caruso, C.M.; Clegg, S.M.; Coulson, T.; DiBattista, J.; Gotanda, K.M.; Francis, C.D.; et al. Precipitation drives global variation in natural selection. Science 2017, 355, 959–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naesset, E.; Orka, H.O.; Solberg, S.; Bollandsas, O.M.; Hansen, E.H.; Mauya, E.; Zahabu, E.; Malimbwi, R.; Chamuya, N.; Olsson, H.; et al. Mapping and estimating forest area and aboveground biomass in miombo woodlands in Tanzania using data from airborne laser scanning, TanDEM-X, RapidEye, and global forest maps: A comparison of estimated precision. Remote Sens. Environ. 2016, 175, 282–300. [Google Scholar] [CrossRef]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote Sensing Technologies for Enhancing Forest Inventories: A Review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef] [Green Version]
- Wulder, M.A.; White, J.C.; Nelson, R.F.; Naesset, E.; Orka, H.O.; Coops, N.C.; Hilker, T.; Bater, C.W.; Gobakken, T. Lidar sampling for large-area forest characterization: A review. Remote Sens. Environ. 2012, 121, 196–209. [Google Scholar] [CrossRef] [Green Version]
- Korpela, I.; Dahlin, B.; Schäfer, H.; Bruun, E.; Haapaniemi, F.; Honkasalo, J.; Ilvesniemi, S.; Kuutti, V.; Linkosalmi, M.; Mustonen, J. Single-tree forest inventory using lidar and aerial images for 3D treetop positioning, species recognition, height and crown width estimation. In Proceedings of the ISPRS Workshop on Laser Scanning and SilviLaser, Espoo, Finland, 12–14 September 2007; IAPRS, 2007; Volume XXXVI, Part 3/W52. pp. 227–233. [Google Scholar]
- Mei, C.; Durrieu, S. Tree crown delineation from digital elevation models and high resolution imagery. International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences. Dir. Open Access J. 2004, 36, 3–6. [Google Scholar]
- Zhao, K.; Suarez, J.C.; Garcia, M.; Hu, T.; Wang, C.; Londo, A. Utility of multitemporal lidar for forest and carbon monitoring: Tree growth, biomass dynamics, and carbon flux. Remote Sens. Environ. 2018, 204, 883–897. [Google Scholar] [CrossRef]
- Nuijten, R.J.; Coops, N.C.; Goodbody, T.R.; Pelletier, G. Examining the Multi-Seasonal Consistency of Individual Tree Segmentation on Deciduous Stands Using Digital Aerial Photogrammetry (DAP) and Unmanned Aerial Systems (UAS). Remote Sens. 2019, 11, 739. [Google Scholar] [CrossRef] [Green Version]
- Lefsky, M.A.; Cohen, W.B.; Parker, G.G.; Harding, D.J. Lidar remote sensing for ecosystem studies. Bioscience 2002, 52, 19–30. [Google Scholar] [CrossRef]
- Chen, Q. Airborne lidar data processing and information extraction. Photogramm. Eng. Remote Sens. 2007, 73, 109. [Google Scholar]
- Csillik, O.; Cherbini, J.; Johnson, R.; Lyons, A.; Kelly, M. Identification of citrus trees from unmanned aerial vehicle imagery using convolutional neural networks. Drones 2018, 2, 39. [Google Scholar] [CrossRef] [Green Version]
- Jennings, S.B.; Brown, N.D.; Sheil, D. Assessing forest canopies and understorey illumination: Canopy closure, canopy cover and other measures. Forestry 1999, 72, 59–73. [Google Scholar] [CrossRef]
- Kelly, M.; Di Tommaso, S. Mapping forests with Lidar provides flexible, accurate data with many uses. Calif. Agric. 2015, 69, 14–20. [Google Scholar] [CrossRef]
- Koch, B.; Heyder, U.; Weinacker, H. Detection of individual tree crowns in airborne lidar data. Photogramm. Eng. Remote Sens. 2006, 72, 357–363. [Google Scholar] [CrossRef] [Green Version]
- Jakubowski, M.; Li, W.; Guo, Q.; Kelly, M. Delineating individual trees from LiDAR data: A comparison of vector-and raster-based segmentation approaches. Remote Sens. 2013, 5, 4163–4186. [Google Scholar] [CrossRef] [Green Version]
- Jing, L.; Hu, B.; Li, J.; Noland, T. Automated delineation of individual tree crowns from LiDAR data by multi-scale analysis and segmentation. Photogramm. Eng. Remote Sens. 2012, 78, 1275–1284. [Google Scholar] [CrossRef]
- Kaartinen, H.; Hyyppä, J.; Yu, X.; Vastaranta, M.; Hyyppä, H.; Kukko, A.; Holopainen, M.; Heipke, C.; Hirschmugl, M.; Morsdorf, F. An international comparison of individual tree detection and extraction using airborne laser scanning. Remote Sens. 2012, 4, 950–974. [Google Scholar] [CrossRef] [Green Version]
- Hyyppa, J.; Kelle, O.; Lehikoinen, M.; Inkinen, M. A segmentation-based method to retrieve stem volume estimates from 3-D tree height models produced by laser scanners. IEEE Trans. Geosci. Remote Sens. 2001, 39, 969–975. [Google Scholar] [CrossRef]
- Khosravipour, A.; Skidmore, A.K.; Isenburg, M.; Wang, T.; Hussin, Y.A. Generating Pit-free Canopy Height Models from Airborne Lidar. Photogramm. Eng. Remote Sens. 2014, 80, 863–872. [Google Scholar] [CrossRef]
- Lu, X.; Guo, Q.; Li, W.; Flanagan, J. A bottom-up approach to segment individual deciduous trees using leaf-off lidar point cloud data. ISPRS J. Photogramm. Remote Sens. 2014, 94, 1–12. [Google Scholar] [CrossRef]
- Chen, Q.; Baldocchi, D.; Gong, P.; Kelly, M. Isolating individual trees in a savanna woodland using small footprint lidar data. Photogramm. Eng. Remote Sens. 2006, 72, 923–932. [Google Scholar] [CrossRef] [Green Version]
- Smits, I.; Prieditis, G.; Dagis, S.; Dubrovskis, D. Individual tree identification using different LIDAR and optical imagery data processing methods. Biosyst. Inf. Technol. 2012, 1, 19–24. [Google Scholar] [CrossRef]
- Reitberger, J.; Schnörr, C.; Krzystek, P.; Stilla, U. 3D segmentation of single trees exploiting full waveform LIDAR data. ISPRS J. Photogramm. Remote Sens. 2009, 64, 561–574. [Google Scholar] [CrossRef]
- Li, W.; Guo, Q.; Jakubowski, M.K.; Kelly, M. A New Method for Segmenting Individual Trees from the Lidar Point Cloud. Photogramm. Eng. Remote Sens. 2012, 78, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Hamraz, H.; Contreras, M.A.; Zhang, J. Vertical stratification of forest canopy for segmentation of understory trees within small-footprint airborne LiDAR point clouds. ISPRS J. Photogramm. Remote Sens. 2017, 130, 385–392. [Google Scholar] [CrossRef] [Green Version]
- Ayrey, E.; Fraver, S.; Kershaw, J.A., Jr.; Kenefic, L.S.; Hayes, D.; Weiskittel, A.R.; Roth, B.E. Layer Stacking: A Novel Algorithm for Individual Forest Tree Segmentation from LiDAR Point Clouds. Can. J. Remote Sens. 2017, 43, 16–27. [Google Scholar] [CrossRef]
- Duncanson, L.I.; Cook, B.D.; Hurtt, G.C.; Dubayah, R.O. An efficient, multi-layered crown delineation algorithm for mapping individual tree structure across multiple ecosystems. Remote Sens. Environ. 2014, 154, 378–386. [Google Scholar] [CrossRef]
- Tochon, G.; Feret, J.B.; Valero, S.; Martin, R.E.; Knapp, D.E.; Salembier, P.; Chanussot, J.; Asner, G.P. On the use of binary partition trees for the tree crown segmentation of tropical rainforest hyperspectral images. Remote Sens. Environ. 2015, 159, 318–331. [Google Scholar] [CrossRef] [Green Version]
- Paris, C.; Valduga, D.; Bruzzone, L. A hierarchical approach to three-dimensional segmentation of LiDAR data at single-tree level in a multilayered forest. IEEE Trans. Geosci. Remote Sens. 2016, 54, 4190–4203. [Google Scholar] [CrossRef]
- Leckie, D.; Gougeon, F.; Hill, D.; Quinn, R.; Armstrong, L.; Shreenan, R. Combined high-density lidar and multispectral imagery for individual tree crown analysis. Can. J. Remote Sens. 2003, 29, 633–649. [Google Scholar] [CrossRef]
- Pouliot, D.A.; King, D.J.; Pitt, D.G. Development and evaluation of an automated tree detection-delineation algorithm for monitoring regenerating coniferous forests. Can. J. For. Res. Rev. Can. Rech. For. 2005, 35, 2332–2345. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Hyyppä, J.; Liang, X.; Kaartinen, H.; Yu, X.; Lindberg, E.; Holmgren, J.; Qin, Y.; Mallet, C.; Ferraz, A. International benchmarking of the individual tree detection methods for modeling 3-D canopy structure for silviculture and forest ecology using airborne laser scanning. IEEE Trans. Geosci. Remote Sens. 2016, 54, 5011–5027. [Google Scholar] [CrossRef] [Green Version]
- Morsdorf, F.; Meier, E.; Kotz, B.; Itten, K.I.; Dobbertin, M.; Allgower, B. LIDAR-based geometric reconstruction of boreal type forest stands at single tree level for forest and wildland fire management. Remote Sens. Environ. 2004, 92, 353–362. [Google Scholar] [CrossRef]
- Sokolova, M.; Japkowicz, N.; Szpakowicz, S. Beyond accuracy, F-Score and ROC: A family of discriminant measures for performance evaluation. In Proceedings of the Ai 2006: Advances in Artificial Intelligence, Hobart, Australia, 4–8 December 2006; Sattar, A., Kang, B.H., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4304, pp. 1015–1021. [Google Scholar]
- Peuhkurinen, J.; Mehtätalo, L.; Maltamo, M. Comparing individual tree detection and the area-based statistical approach for the retrieval of forest stand characteristics using airborne laser scanning in Scots pine stands. Can. J. For. Res. 2011, 41, 583–598. [Google Scholar] [CrossRef]
- Falkowski, M.J.; Smith, A.M.S.; Gessler, P.E.; Hudak, A.T.; Vierling, L.A.; Evans, J.S. The influence of conifer forest canopy cover on the accuracy of two individual tree measurement algorithms using lidar data. Can. J. Remote Sens. 2008, 34, S338–S350. [Google Scholar] [CrossRef]
- Vauhkonen, J.; Ene, L.; Gupta, S.; Heinzel, J.; Holmgren, J.; Pitkanen, J.; Solberg, S.; Wang, Y.; Weinacker, H.; Hauglin, K.M.; et al. Comparative testing of single-tree detection algorithms under different types of forest. Forestry 2012, 85, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Forzieri, G.; Guarnieri, L.; Vivoni, E.R.; Castelli, F.; Preti, F. Multiple attribute decision making for individual tree detection using high-resolution laser scanning. For. Ecol. Manag. 2009, 258, 2501–2510. [Google Scholar] [CrossRef]
- Flood, M. LiDAR activities and research priorities in the commercial sector. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2001, 34, 3–8. [Google Scholar]
- Smith, A.M.S.; Falkowski, M.J.; Hudak, A.T.; Evans, J.S.; Robinson, A.P.; Steele, C.M. A cross-comparison of field, spectral, and lidar estimates of forest canopy cover. Can. J. Remote Sens. 2009, 35, 447–459. [Google Scholar] [CrossRef]
- Isenburg, M. LAStools: Software for rapid LiDAR processing. Plaça Ferrater Mora 2014, 1, 17071. Available online: http://www. liblas. org/ (accessed on 23 August 2019).
- Zhao, X.; Su, Y.; Li, W.; Hu, T.; Liu, J.; Guo, Q. A Comparison of LiDAR Filtering Algorithms in Vegetated Mountain Areas. Can. J. Remote Sens. 2018, 44, 287–298. [Google Scholar] [CrossRef]
- Axelsson, P. Processing of laser scanner data—Algorithms and applications. ISPRS J. Photogramm. Remote Sens. 1999, 54, 138–147. [Google Scholar] [CrossRef]
- Meng, X.; Currit, N.; Zhao, K. Ground filtering algorithms for airborne LiDAR data: A review of critical issues. Remote Sens. 2010, 2, 833–860. [Google Scholar] [CrossRef] [Green Version]
- Guo, Q.; Li, W.; Yu, H.; Alvarez, O. Effects of Topographic Variability and Lidar Sampling Density on Several DEM Interpolation Methods. Photogramm. Eng. Remote Sens. 2010, 76, 701–712. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Slatton, K.C.; Roth, B.E.; Cropper, W.P., Jr. Adaptive clustering of airborne LiDAR data to segment individual tree crowns in managed pine forests. Int. J. Remote Sens. 2010, 31, 117–139. [Google Scholar] [CrossRef]
- Nyquist, H. Certain topics in telegraph transmission theory. Trans. Am. Inst. Electr. Eng. 1928, 47, 617–644. [Google Scholar] [CrossRef]
- Soille, P. Morphological Image Analysis: Principles and Applications; Springer Science & Business Media: Berlin, Germany, 2013. [Google Scholar]
- Pitkänen, J.; Maltamo, M.; Hyyppä, J.; Yu, X. Adaptive methods for individual tree detection on airborne laser based canopy height model. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2004, 36, 187–191. [Google Scholar]
- Goutte, C.; Gaussier, E. A probabilistic interpretation of precision, recall and F-score, with implication for evaluation. In Advances in Information Retrieval, Proceedings of the European Conference on Information Retrieval, Spain, 21–23 March 2005; Losada, D.E., FernandezLuna, J.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; Volume 3408, pp. 345–359. [Google Scholar]
- Chen, J.M.; Pavlic, G.; Brown, L.; Cihlar, J.; Leblanc, S.; White, H.; Hall, R.; Peddle, D.; King, D.; Trofymow, J. Derivation and validation of Canada-wide coarse-resolution leaf area index maps using high-resolution satellite imagery and ground measurements. Remote Sens. Environ. 2002, 80, 165–184. [Google Scholar] [CrossRef]
- Ma, Q.; Su, Y.; Guo, Q. Comparison of Canopy Cover Estimations From Airborne LiDAR, Aerial Imagery, and Satellite Imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 4225–4236. [Google Scholar] [CrossRef]
- Johnson, J.W. A heuristic method for estimating the relative weight of predictor variables in multiple regression. Multivar. Behav. Res. 2000, 35, 1–19. [Google Scholar] [CrossRef]
- Dralle, K.; Rudemo, M. Stem number estimation by kernel smoothing of aerial photos. Can. J. For. Res. 1996, 26, 1228–1236. [Google Scholar] [CrossRef]
- Persson, A.; Holmgren, J.; Soderman, U. Detecting and measuring individual trees using an airborne laser scanner. Photogramm. Eng. Remote Sens. 2002, 68, 925–932. [Google Scholar]
- Magnussen, S.; Eggermont, P.; LaRiccia, V.N. Recovering Tree Heights from Airborne Laser Scanner Data. For. Sci. 1999, 45, 407–422. [Google Scholar]
- Zheng, G.; Moskal, L.M. Computational-Geometry-Based Retrieval of Effective Leaf Area Index Using Terrestrial Laser Scanning. IEEE Trans. Geosci. Remote Sens. 2012, 50, 3958–3969. [Google Scholar] [CrossRef]
- Maltamo, M.; Mustonen, K.; Hyyppä, J.; Pitkänen, J.; Yu, X. The accuracy of estimating individual tree variables with airborne laser scanning in a boreal nature reserve. Can. J. For. Res. 2004, 34, 1791–1801. [Google Scholar] [CrossRef]
- Falkowski, M.J.; Smith, A.M.S.; Hudak, A.T.; Gessler, P.E.; Vierling, L.A.; Crookston, N.L. Automated estimation of individual conifer tree height and crown diameter via two-dimensional spatial wavelet analysis of lidar data. Can. J. Remote Sens. 2006, 32, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Zawawi, A.A.; Shiba, M.; Jemali, N.J.N. Accuracy of LiDAR-based tree height estimation and crown recognition in a subtropical evergreen broad-leaved forest in Okinawa, Japan. For. Syst. 2015, 24, 1–11. [Google Scholar] [CrossRef]
- Magnussen, S.; Boudewyn, P. Derivations of stand heights from airborne laser scanner data with canopy-based quantile estimators. Can. J. For. Res. 1998, 28, 1016–1031. [Google Scholar] [CrossRef]
- Ozdemir, I.; Norton, D.A.; Ozkan, U.Y.; Mert, A.; Senturk, O. Estimation of Tree Size Diversity Using Object Oriented Texture Analysis and Aster Imagery. Sensors (Basel) 2008, 8, 4709–4724. [Google Scholar] [CrossRef] [Green Version]
- Alteyrac, J.; Cloutier, A.; Zhang, S.Y. Characterization of juvenile wood to mature wood transition age in black spruce (Picea mariana (Mill.) BSP) at different stand densities and sampling heights. Wood Sci. Technol. 2006, 40, 124–138. [Google Scholar] [CrossRef]
- Jung, S.E.; Kwak, D.A.; Park, T.; Lee, W.K.; Yoo, S. Estimating Crown Variables of Individual Trees Using Airborne and Terrestrial Laser Scanners. Remote Sens. 2011, 3, 2346–2363. [Google Scholar] [CrossRef] [Green Version]
- Stonge, B.A.; Cavayas, F. Estimating forest stand structure from high resolution imagery using the directional variogram. Int. J. Remote Sens. 1995, 16, 1999–2021. [Google Scholar] [CrossRef]
- Graves, S.J.; Caughlin, T.T.; Asner, G.P.; Bohlman, S.A. A tree-based approach to biomass estimation from remote sensing data in a tropical agricultural landscape. Remote Sens. Environ. 2018, 218, 32–43. [Google Scholar] [CrossRef]
- García, M.; Gajardo, J.; Riaño, D.; Zhao, K.; Martín, P.; Ustin, S. Canopy clumping appraisal using terrestrial and airborne laser scanning. Remote Sens. Environ. 2015, 161, 78–88. [Google Scholar] [CrossRef]
- Mehtätalo, L. Eliminating the effect of overlapping crowns from aerial inventory estimates. Can. J. For. Res. 2006, 36, 1649–1660. [Google Scholar] [CrossRef]








| Algorithm | Reference | Vegetation Type | Point Density (pts/m2) | Accuracy (%) | Evaluation Method |
|---|---|---|---|---|---|
| Region growing a | Hyyppä et al. (2001) [19] | Conifer | 8–10 | - | - |
| Pouring a | Koch et al. (2006) [15] | Conifer, Broadleaf | 5/10 | 62 | MARA |
| Watershed a | Jing et al. (2012) [17] | Conifer, Broadleaf | 45 | 69 | C, O |
| Marker-controlled Watershed a | Chen et al. (2006) [22] | Conifer | 9.5 | 64 | AATI |
| Local maxima a | Smits et al. (2012) [23] | Conifer, Broadleaf | 9 | 87.50 | D |
| Pit-free canopy height model/PFCHM a | Khosravipour et al. (2014) [20] | Conifer, Broadleaf | 160 | 74 | AI |
| Normalized cut b | Reitberger et al. (2009) [24] | Conifer, Broadleaf | 25/10 | 66 | - |
| Point cloud segmentation b | Li et al. (2012) [25] | Conifer | 6 | 90 | F-score |
| Bottom-up region growing b | Lu et al. (2014) [21] | Broadleaf | 10.28 | 84 | F-score |
| Region growing b | Hamraz et al. (2017) [26] | Broadleaf | 25/1.5 | - | - |
| Layer stacking b | Ayrey et al. (2017) [27] | Conifer, Broadleaf | 21/6/5 | 72 | D, C, O |
| Iterative watershed b | Duncanson et al. (2014) [28] | Broadleaf | 18 | 70 | - |
| Watershed + k-means b | Tochon et al. (2015) [29] | Conifer, Broadleaf | - | 69.86 | D, US, M, OS |
| Hierarchical approach b | Paris et al. (2016) [30] | Conifer (Multilayer) | 50/8 | 92–97 | D, C, O |
| Forest Type | Leaf Area Index (LAI) | Canopy Cover (%) | Tree Density (Trees/ha) | Coefficient of Variation of Tree Height (CVTH) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Maximum | Minimum | Mean | SD | Maximum | Minimum | Mean | SD | Maximum | Minimum | Mean | SD | Maximum | Minimum | Mean | SD | |
| Conifer | 2.41 | 0.14 | 1.25 | 0.54 | 0.79 | 0.07 | 0.60 | 0.18 | 520 | 100 | 314 | 113 | 1.19 | 0.21 | 0.47 | 0.17 |
| Broadleaf | 4.59 | 0.32 | 2.66 | 0.44 | 0.96 | 0.45 | 0.82 | 0.07 | 439 | 85 | 278 | 73 | 0.70 | 0.06 | 0.23 | 0.14 |
| Mixed | 4.51 | 0.61 | 3.40 | 0.38 | 0.97 | 0.69 | 0.90 | 0.03 | 538 | 255 | 397 | 74 | 0.50 | 0.05 | 0.18 | 0.11 |
| Forest Type | LAI | Canopy Cover (%) | Tree Density (Trees/ha) | CVTH | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Level 1 | Level 2 | Level 3 | Level 1 | Level 2 | Level 3 | Level 1 | Level 2 | Level 3 | Level 1 | Level 2 | Level 3 | |
| Conifer | <1 | 1–1.7 | >1.7 | <0.3 | 0.3–0.55 | >0.55 | <240 | 240–360 | >360 | 0.5 | 0.5–0.8 | 0.8 |
| Broadleaf | <2 | 2–3.2 | >3.2 | <0.65 | 0.65–0.85 | >0.85 | <226 | 226–325 | >325 | 0.2 | 0.2–0.3 | 0.3 |
| Mixed | <2.7 | 2.7–3.6 | >3.6 | <0.8 | 0.8–0.9 | >0.9 | <254 | 254–439 | >439 | 0.2 | 0.2–0.3 | 0.3 |
| Forest Type | CHM | PFCHM | PCS | LSS | ||||
|---|---|---|---|---|---|---|---|---|
| F-Score | OA | F-Score | OA | F-Score | OA | F-Score | OA | |
| Conifer | 0.80 | 0.82 | 0.88 | 0.90 | 0.82 | 0.83 | 0.84 | 0.86 |
| Broadleaf | 0.78 | 0.78 | 0.80 | 0.83 | 0.68 | 0.79 | 0.76 | 0.80 |
| Mixed | 0.79 | 0.79 | 0.85 | 0.87 | 0.77 | 0.82 | 0.80 | 0.83 |
| Forest Type | CHM | PFCHM | PCS | LSS |
|---|---|---|---|---|
| Conifer | 0.30 | 0.20 | 0.13 | 0.34 |
| Broadleaf | 0.19 | 0.07 | 0.49 | 0.36 |
| Mixed | 0.48 | 0.56 | 0.50 | 0.29 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Q.; Su, Y.; Jin, S.; Kelly, M.; Hu, T.; Ma, Q.; Li, Y.; Song, S.; Zhang, J.; Xu, G.; et al. The Influence of Vegetation Characteristics on Individual Tree Segmentation Methods with Airborne LiDAR Data. Remote Sens. 2019, 11, 2880. https://doi.org/10.3390/rs11232880
Yang Q, Su Y, Jin S, Kelly M, Hu T, Ma Q, Li Y, Song S, Zhang J, Xu G, et al. The Influence of Vegetation Characteristics on Individual Tree Segmentation Methods with Airborne LiDAR Data. Remote Sensing. 2019; 11(23):2880. https://doi.org/10.3390/rs11232880
Chicago/Turabian StyleYang, Qiuli, Yanjun Su, Shichao Jin, Maggi Kelly, Tianyu Hu, Qin Ma, Yumei Li, Shilin Song, Jing Zhang, Guangcai Xu, and et al. 2019. "The Influence of Vegetation Characteristics on Individual Tree Segmentation Methods with Airborne LiDAR Data" Remote Sensing 11, no. 23: 2880. https://doi.org/10.3390/rs11232880