National Scale 3D Mapping of Soil pH Using a Data Augmentation Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Data
2.2.1. Soil pH Data
2.2.2. Environmental Covariates
2.3. Depth as a Covariate Using Data Augmentation
2.3.1. Augmenting Depth Information
2.3.2. Augmenting Attribute Information
2.4. Modelling Framework
2.4.1. Variable Selection
2.4.2. Calibration of the Soil pH Model
2.5. Evaluation and Comparison to Existing Products
2.6. Adaptation of the Predictions to Suit Different End-Users
3. Results
3.1. Soil Data
3.2. Selected Predictors and Variable Importance
3.3. Impact of Data Augmentation on Predictions
3.4. Model Evaluation
3.5. Generated Products
3.5.1. Depth Profiles
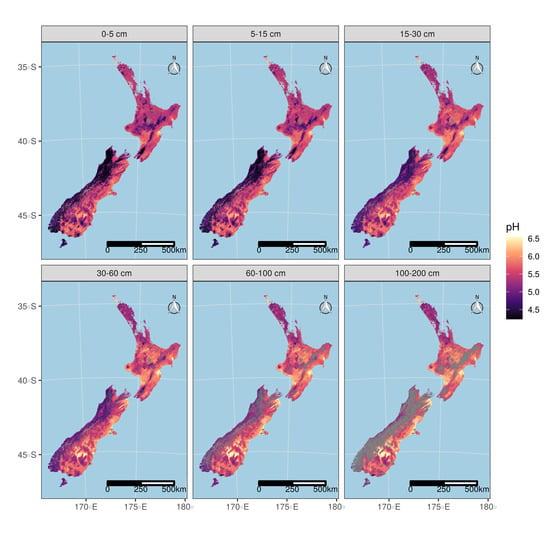
3.5.2. Maps
3.6. Uncertainty of the Predictions Across the Country
4. Discussion
4.1. Comparison with 2.5D DSM
4.2. Comparison with Other Soil pH Products for New Zealand
4.3. Limitations and Potential Improvements
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sparks, D.L. Environmental Soil Chemistry; Elsevier: Amsterdam, The Netherlands, 2003. [Google Scholar]
- Neina, D. The role of soil pH in plant nutrition and soil remediation. Appl. Environ. Soil Sci. 2019, 2019, 5794869. [Google Scholar] [CrossRef]
- Cavanagh, J.A.E.; Yi, Z.; Gray, C.; Munir, K.; Lehto, N.; Robinson, B. Cadmium uptake by onions, lettuce and spinach in New Zealand: Implications for management to meet regulatory limits. Sci. Total Environ. 2019, 668, 780–789. [Google Scholar] [CrossRef]
- Johnson, P.; Gerbeaux, P. Wetland Types in New Zealand; Department of Conservation: Wellington, New Zealand, 2004. [Google Scholar]
- Webb, T.; Wilson, A. A Manual of Land Characteristics for Evaluation of Rural Land; Landcare Research Science Series 10; Manaaki Whenua Press: Lincoln, New Zealand, 1995. [Google Scholar]
- Ausseil, A.G.; Lindsay Chadderton, W.; Gerbeaux, P.; Theo Stephens, R.; Leathwick, J.R. Applying systematic conservation planning principles to palustrine and inland saline wetlands of New Zealand. Freshw. Biol. 2011, 56, 142–161. [Google Scholar] [CrossRef]
- Ringrose-Voase, A.; Grealish, G.; Thomas, M.; Wong, M.; Glover, M.; Mercado, A.; Nilo, G.; Dowling, T. Four Pillars of digital land resource mapping to address information and capacity shortages in developing countries. Geoderma 2019, 352, 299–313. [Google Scholar] [CrossRef]
- Kidd, D.; Webb, M.; Malone, B.; Minasny, B.; McBratney, A.B. Digital soil assessment of agricultural suitability, versatility and capital in Tasmania, Australia. Geoderma Reg. 2015, 6, 7–21. [Google Scholar] [CrossRef]
- Sparling, G.; Schipper, L. Soil quality at a national scale in New Zealand. J. Environ. Qual. 2002, 31, 1848–1857. [Google Scholar] [CrossRef]
- Ministry for the Environment; Statistics New Zealand. New Zealand’s Environmental Reporting Series: Our Land 2018; Ministry for the Environment: Wellington, New Zealand; Statistics New Zealand: Wellington, New Zealand, 2018.
- Kidd, D.; Field, D.; McBratney, A.B.; Webb, M. A preliminary spatial quantification of the soil security dimensions for Tasmania. Geoderma 2018, 322, 184–200. [Google Scholar] [CrossRef]
- Landcare Research. New Zealand Fundamental Soil Layers, Soil pH. 2000. Available online: https://lris.scinfo.org.nz/layer/48102-fsl-ph/ (accessed on 15 July 2020).
- Hengl, T.; de Jesus, J.M.; Heuvelink, G.B.; Gonzalez, M.R.; Kilibarda, M.; Blagotić, A.; Shangguan, W.; Wright, M.N.; Geng, X.; Bauer-Marschallinger, B.; et al. SoilGrids250m: Global gridded soil information based on machine learning. PLoS ONE 2017, 12, e0169748. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Mulder, V.L.; Heuvelink, G.B.; Poggio, L.; Caubet, M.; Dobarco, M.R.; Walter, C.; Arrouays, D. Model averaging for mapping topsoil organic carbon in France. Geoderma 2020, 366, 114237. [Google Scholar] [CrossRef]
- Arrouays, D.; Leenaars, J.G.; Richer-de Forges, A.C.; Adhikari, K.; Ballabio, C.; Greve, M.; Grundy, M.; Guerrero, E.; Hempel, J.; Hengl, T.; et al. Soil legacy data rescue via GlobalSoilMap and other international and national initiatives. GeoResJ 2017, 14, 1–19. [Google Scholar] [CrossRef]
- Bishop, T.; McBratney, A.; Laslett, G. Modelling soil attribute depth functions with equal-area quadratic smoothing splines. Geoderma 1999, 91, 27–45. [Google Scholar] [CrossRef]
- Zhang, G.l.; Feng, L.; Song, X.D. Recent progress and future prospect of digital soil mapping: A review. J. Integr. Agric. 2017, 16, 2871–2885. [Google Scholar] [CrossRef]
- Jenny, H. Factors of Soil Formation: A System of Quantitative Pedology; McGraw-Hill: New York, NY, USA, 1941. [Google Scholar]
- Minasny, B.; McBratney, A.B.; Mendonça-Santos, M.; Odeh, I.; Guyon, B. Prediction and digital mapping of soil carbon storage in the Lower Namoi Valley. Soil Res. 2006, 44, 233–244. [Google Scholar] [CrossRef]
- Myers, D.B.; Kitchen, N.R.; Sudduth, K.A.; Miles, R.J.; Sadler, E.J.; Grunwald, S. Peak functions for modeling high resolution soil profile data. Geoderma 2011, 166, 74–83. [Google Scholar] [CrossRef]
- Malone, B.P.; McBratney, A.; Minasny, B.; Laslett, G. Mapping continuous depth functions of soil carbon storage and available water capacity. Geoderma 2009, 154, 138–152. [Google Scholar] [CrossRef]
- Orton, T.; Pringle, M.; Bishop, T. A one-step approach for modelling and mapping soil properties based on profile data sampled over varying depth intervals. Geoderma 2016, 262, 174–186. [Google Scholar] [CrossRef]
- Pejović, M.; Nikolić, M.; Heuvelink, G.B.; Hengl, T.; Kilibarda, M.; Bajat, B. Sparse regression interaction models for spatial prediction of soil properties in 3D. Comput. Geosci. 2018, 118, 1–13. [Google Scholar] [CrossRef]
- Ramcharan, A.; Hengl, T.; Nauman, T.; Brungard, C.; Waltman, S.; Wills, S.; Thompson, J. Soil property and class maps of the conterminous United States at 100-meter spatial resolution. Soil Sci. Soc. Am. J. 2018, 82, 186–201. [Google Scholar] [CrossRef] [Green Version]
- Brus, D.; Yang, R.M.; Zhang, G.L. Three-dimensional geostatistical modeling of soil organic carbon: A case study in the Qilian Mountains, China. Catena 2016, 141, 46–55. [Google Scholar] [CrossRef]
- Nauman, T.W.; Duniway, M.C. Relative prediction intervals reveal larger uncertainty in 3D approaches to predictive digital soil mapping of soil properties with legacy data. Geoderma 2019, 347, 170–184. [Google Scholar] [CrossRef]
- Minasny, B.; McBratney, A.B. Digital soil mapping: A brief history and some lessons. Geoderma 2016, 264, 301–311. [Google Scholar] [CrossRef]
- Manaaki Whenua—Landcare Research. National Soils Data Repository. 2020. Available online: https://doi.org/10.26060/95m4-cz25 (accessed on 15 July 2020).
- Libohova, Z.; Wills, S.; Odgers, N.P.; Ferguson, R.; Nesser, R.; Thompson, J.A.; West, L.T.; Hempel, J.W. Converting pH 1:1 H2O and 1:2 CaCl2 to 1:5 H2O to contribute to a harmonized global soil database. Geoderma 2014, 213, 544–550. [Google Scholar] [CrossRef]
- Holdaway, R.J.; Easdale, T.A.; Carswell, F.E.; Richardson, S.J.; Peltzer, D.A.; Mason, N.W.; Brandon, A.M.; Coomes, D.A. Nationally representative plot network reveals contrasting drivers of net biomass change in secondary and old-growth forests. Ecosystems 2017, 20, 944–959. [Google Scholar] [CrossRef] [Green Version]
- Grafström, A.; Lundström, N.L.; Schelin, L. Spatially balanced sampling through the pivotal method. Biometrics 2012, 68, 514–520. [Google Scholar] [CrossRef]
- Grafström, A.; Lisic, J. BalancedSampling: Balanced and Spatially Balanced Sampling, R Package Version 1.5.5; 2019. Available online: https://CRAN.R-project.org/package=BalancedSampling (accessed on 15 July 2020).
- Olaya, V.; Conrad, O. Geomorphometry in SAGA. Dev. Soil Sci. 2009, 33, 293–308. [Google Scholar]
- Neteler, M.; Mitasova, H. Open Source GIS: A GRASS GIS Approach; Springer Science Business Media: Berlin/Heidelberg, Germany, 2013; Volume 689. [Google Scholar]
- Running, S.W.; Thornton, P.E.; Nemani, R.; Glassy, J.M. Global terrestrial gross and net primary productivity from the earth observing system. In Methods in Ecosystem Science; Springer: Berlin/Heidelberg, Germany, 2000; pp. 44–57. [Google Scholar]
- Leathwick, J.; Morgan, F.; Wilson, G.; Rutledge, D.; McLeod, M.; Johnston, K. Land Environments of New Zealand: A Technical Guide; Manaaki Whenua Press: Lincoln, New Zealand, 2002; p. 184. [Google Scholar]
- Tait, A.; Henderson, R.; Turner, R.; Zheng, X. Thin plate smoothing spline interpolation of daily rainfall for New Zealand using a climatological rainfall surface. Int. J. Climatol. 2006, 26, 2097–2115. [Google Scholar] [CrossRef]
- Porteous, A.; Basher, R.; Salinger, M. Calibration and performance of the single-layer soil water balance model for pasture sites. N. Z. J. Agric. Res. 1994, 37, 107–118. [Google Scholar] [CrossRef]
- Hofierka, J.; Suri, M. The solar radiation model for Open source GIS: Implementation and applications. In Proceedings of the Open Source GIS-GRASS Users Conference, Trento, Italy, 11–13 September 2002; pp. 1–19. [Google Scholar]
- Landcare Research. LCDB v4.1—Landcover Database Version 4.1. 2015. Available online: https://lris.scinfo.org.nz/layer/48423-lcdb-v41-land-cover-database-version-41-mainland-new-zealand/ (accessed on 15 July 2020).
- Leathwick, J. New Zealand’s potential forest pattern as predicted from current species-environment relationships. N. Z. J. Bot. 2001, 39, 447–464. [Google Scholar] [CrossRef] [Green Version]
- Landcare Research. NZDEM—New Zealand Digital Elevation Model. 2010. Available online: https://lris.scinfo.org.nz/layer/48131-nzdem-north-island-25-metre/ (accessed on 15 July 2020).
- Available online: https://lris.scinfo.org.nz/layer/48127-nzdem-south-island-25-metre/ (accessed on 15 July 2020).
- Landcare Research. New Zealand Land Resource Inventory. 2010. Available online: https://lris.scinfo.org.nz/layer/48065-nzlri-rock/ (accessed on 15 July 2020).
- Minasny, B.; Berglund, Ö.; Connolly, J.; Hedley, C.; de Vries, F.; Gimona, A.; Kempen, B.; Kidd, D.; Lilja, H.; Malone, B.; et al. Digital mapping of peatlands—A critical review. Earth Sci. Rev. 2019, 196, 102870. [Google Scholar] [CrossRef]
- Tanner, M.A.; Wong, W.H. The calculation of posterior distributions by data augmentation. J. Am. Stat. Assoc. 1987, 82, 528–540. [Google Scholar] [CrossRef]
- Cubuk, E.D.; Zoph, B.; Mane, D.; Vasudevan, V.; Le Quoc, V. AutoAugment: Learning augmentation strategies from data. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15 June 2019; pp. 113–123. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Probst, P.; Wright, M.N.; Boulesteix, A.L. Hyperparameters and tuning strategies for random forest. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9, e1301. [Google Scholar] [CrossRef] [Green Version]
- Wright, M.N.; Ziegler, A. ranger: A fast implementation of random forests for high dimensional data in C++ and R. J. Stat. Softw. 2017, 77, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Meinshausen, N. Quantile regression forests. J. Mach. Learn. Res. 2006, 7, 983–999. [Google Scholar]
- Vaysse, K.; Lagacherie, P. Using quantile regression forest to estimate uncertainty of digital soil mapping products. Geoderma 2017, 291, 55–64. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Xiong, X.; Grunwald, S.; Myers, D.B.; Kim, J.; Harris, W.G.; Comerford, N.B. Holistic environmental soil-landscape modeling of soil organic carbon. Environ. Model. Softw. 2014, 57, 202–215. [Google Scholar] [CrossRef]
- Meyer, H.; Reudenbach, C.; Wöllauer, S.; Nauss, T. Importance of spatial predictor variable selection in machine learning applications–Moving from data reproduction to spatial prediction. Ecol. Model. 2019, 411, 108815. [Google Scholar] [CrossRef] [Green Version]
- Genuer, R.; Poggi, J.M.; Tuleau-Malot, C. Variable selection using random forests. Pattern Recognit. Lett. 2010, 31, 2225–2236. [Google Scholar] [CrossRef] [Green Version]
- Genuer, R.; Poggi, J.M.; Tuleau-Malot, C. VSURF: Variable Selection Using Random Forests; R Package Version 1.1.0; 2019. Available online: https://CRAN.R-project.org/package=VSURF/ (accessed on 15 July 2020).
- Meyer, H. CAST: ’caret’ Applications for Spatial-Temporal Models; R Package Version 0.3.2; 2019. Available online: https://CRAN.R-project.org/package=CAST/ (accessed on 15 July 2020).
- New Zealand Soil Bureau. Soils of New Zealand, Part 2 (Soil Bureau Bulletin 26 (2)); New Zealand Departement of Scientific and Industrial Research: Wellington, New Zealand, 1968.
- Molloy, L. Soils in the New Zealand Landscape: The Living Mantle, 2nd ed.; Mallinson Rendel Publishers Ltd.: Wellington, New Zealand, 1988. [Google Scholar]
- Beaudette, D.; Roudier, P.; O’Geen, A. Algorithms for quantitative pedology: A toolkit for soil scientists. Comput. Geosci. 2013, 52, 258–268. [Google Scholar] [CrossRef]
- Biro, K.; Pradhan, B.; Buchroithner, M.; Makeschin, F. Land use/land cover change analysis and its impact on soil properties in the northern part of Gadarif region, Sudan. Land Degrad. Dev. 2013, 24, 90–102. [Google Scholar] [CrossRef]
- Slessarev, E.; Lin, Y.; Bingham, N.; Johnson, J.; Dai, Y.; Schimel, J.; Chadwick, O. Water balance creates a threshold in soil pH at the global scale. Nature 2016, 540, 567–569. [Google Scholar] [CrossRef] [PubMed]
- Wadoux, A.M.C.; Samuel-Rosa, A.; Poggio, L.; Mulder, V.L. A note on knowledge discovery and machine learning in digital soil mapping. Eur. J. Soil Sci. 2020, 71, 133–136. [Google Scholar] [CrossRef]
- Pärtel, M. Local plant diversity patterns and evolutionary history at the regional scale. Ecology 2002, 83, 2361–2366. [Google Scholar] [CrossRef]
- Allen, R.; McIntosh, P.; Wilson, J. The distribution of plants in relation to pH and salinity on inland saline/alkaline soils in Central Otago, New Zealand. N. Z. J. Bot. 1997, 35, 517–523. [Google Scholar] [CrossRef]
- Williams, P.A.; Wiser, S.; Clarkson, B.; Stanley, M.C. New Zealand’s historically rare terrestrial ecosystems set in a physical and physiognomic framework. N. Z. J. Ecol. 2007, 31, 119–128. [Google Scholar]
- Viscarra Rossel, R.; Chen, C.; Grundy, M.; Searle, R.; Clifford, D.; Campbell, P. The Australian three-dimensional soil grid: Australia’s contribution to the GlobalSoilMap project. Soil Res. 2015, 53, 845–864. [Google Scholar] [CrossRef] [Green Version]
- Nussbaum, M.; Spiess, K.; Baltensweiler, A.; Grob, U.; Keller, A.; Greiner, L.; Schaepman, M.E.; Papritz, A.J. Evaluation of digital soil mapping approaches with large sets of environmental covariates. Soil 2018, 4, 1–22. [Google Scholar] [CrossRef] [Green Version]











| Name | Original Scale/Resolution | Reference/Formula |
|---|---|---|
| Climate | ||
| Mean annual evapotranspiration 2005–2014 | 500 m | Running et al. [35] |
| Mean annual water deficit 1950–1980 | 25 m | Leathwick et al. [36] |
| Mean annual rainfall 1972–2014 | 5110 m | Tait et al. [37] |
| Mean annual temperature 1950–1980 | 25 m | Leathwick et al. [36] |
| Monthly water balance | 25 m | Leathwick et al. [36] |
| Potential evapotranspiration deficit 1972–2014 | 5096 m | Porteous et al. [38] |
| Annual global solar radiation | 25 m | Neteler and Mitasova [34], Hofierka et al. [39] |
| Organisms | ||
| Mean annual net primary production 2005–2014 | 500 m | Running et al. [35] |
| Land cover (2012) | 1:50,000 | Landcare Research [40] |
| Potential vegetation | 100 m | Leathwick [41] |
| Landscape position | ||
| Elevation | 25 m | Landcare Research [42,43] |
| Aspect | 25 m | Neteler and Mitasova [34] |
| Northerness | 25 m | [34] |
| Easterness | 25 m | Neteler and Mitasova [34] |
| Latitude | 25 m | Neteler and Mitasova [34] |
| Distance from coast | 25 m | Neteler and Mitasova [34] |
| Relief/Topography | ||
| Distance from drainage channels | 25 m | Olaya and Conrad [33] |
| Multi-resolution valley bottom floor (MRVBF) | 25 m | Olaya and Conrad [33] |
| Multi-resolution ridge top floor (MRRTF) | 25 m | Olaya and Conrad [33] |
| Normalised height | 25 m | Olaya and Conrad [33] |
| SAGA wetness index (SWI) | 25 m | Olaya and Conrad [33] |
| Slope | 25 m | Neteler and Mitasova [34] |
| Slope height | 25 m | Olaya and Conrad [33] |
| Standard height | 25 m | Olaya and Conrad [33] |
| Topographic position index (TPI) | 25 m | Olaya and Conrad [33] |
| Valley depth | 25 m | Olaya and Conrad [33] |
| Wind exposition index | 25 m | Olaya and Conrad [33] |
| Underlying geology and erosion | ||
| New Zealand Land Resources Inventory | 1:50,000 | Landcare Research [44] |
| Peat | 1:50,000 | Minasny et al. [45] |
| Depth | N | Min. | Mean | Median | Max. | IQR | Skew. | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0–5 cm | 2,297 | 3.04 | 3.73 | 4.11 | 4.72 | 4.60 | 5.23 | 6.08 | 8.12 | 1.12 | 0.75 |
| 5–15 cm | 2,997 | 3.04 | 3.80 | 4.20 | 4.86 | 4.80 | 5.40 | 6.16 | 8.12 | 1.20 | 0.51 |
| 15–30 cm | 1,660 | 3.30 | 4.30 | 5.20 | 5.53 | 5.60 | 5.91 | 6.46 | 8.30 | 0.71 | −0.02 |
| 30–60 cm | 2,296 | 2.74 | 4.53 | 5.20 | 5.64 | 5.63 | 6.09 | 6.70 | 8.98 | 0.89 | 0.29 |
| 60–100 cm | 1,810 | 2.58 | 4.60 | 5.24 | 5.78 | 5.70 | 6.20 | 7.20 | 9.30 | 0.96 | 0.75 |
| 100–200 cm | 831 | 3.07 | 4.60 | 5.20 | 5.92 | 5.70 | 6.50 | 8.01 | 9.30 | 1.30 | 0.76 |
| 200 cm | 52 | 4.35 | 4.77 | 4.99 | 5.57 | 5.40 | 6.09 | 6.74 | 7.80 | 1.10 | 0.75 |
| All depths | 11,944 | 1.48 | 3.90 | 4.63 | 5.29 | 5.30 | 5.85 | 6.70 | 9.30 | 1.22 | 0.41 |
| RMSE | CCC | Bias | ||
|---|---|---|---|---|
| Our approach | 0.54 | 0.65 | 0.79 | −0.03 |
| 2.5D approach | 0.53 | 0.68 | 0.80 | −0.05 |
| Fundamental Soil Layers | 0.76 | 0.31 | 0.49 | 0.28 |
| SoilGrids 250 m | 0.79 | 0.37 | 0.51 | 0.22 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roudier, P.; Burge, O.R.; Richardson, S.J.; McCarthy, J.K.; Grealish, G.; Ausseil, A.-G. National Scale 3D Mapping of Soil pH Using a Data Augmentation Approach. Remote Sens. 2020, 12, 2872. https://doi.org/10.3390/rs12182872
Roudier P, Burge OR, Richardson SJ, McCarthy JK, Grealish G, Ausseil A-G. National Scale 3D Mapping of Soil pH Using a Data Augmentation Approach. Remote Sensing. 2020; 12(18):2872. https://doi.org/10.3390/rs12182872
Chicago/Turabian StyleRoudier, Pierre, Olivia R. Burge, Sarah J. Richardson, James K. McCarthy, Gerard J. Grealish, and Anne-Gaelle Ausseil. 2020. "National Scale 3D Mapping of Soil pH Using a Data Augmentation Approach" Remote Sensing 12, no. 18: 2872. https://doi.org/10.3390/rs12182872