Detection of European Aspen (Populus tremula L.) Based on an Unmanned Aerial Vehicle Approach in Boreal Forests
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Field Data
2.2. UAV Data
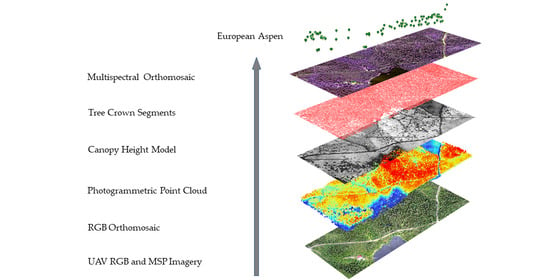
2.3. Calculation of Dense Point Clouds and Image Orthomosaics
2.4. Individual Tree Detection and Spectral Data Features Extraction
2.5. Statistical Analysis
3. Results
3.1. Accuracy Assessment of the 2-Class Classification Model
3.2. Accuracy Assessment of the 4-Class Classification Model
3.3. Spectral Feature Importance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Research Site | Flight | Date | UAV Platform | Sensor | Number of Images | Overlap, % | GSD, cm |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 10.7.18 | eBee plus RTK | S.O.D.A | 341 | 85/85 | 4.7 |
| 2 | 10.7.18 | eBee plus RTK | S.O.D.A | 670 | 85/85 | 4.7 | |
| 3 | 10.7.18 | eBee plus RTK | Sequoia | 864 | 80/80 | 13.7 | |
| 4 | 10.7.18 | eBee plus RTK | Sequoia | 936 | 80/80 | 13.7 | |
| 2 | 5 | 10.7.18 | eBee plus RTK | S.O.D.A | 303 | 85/85 | 4.9 |
| 6 | 10.7.18 | eBee plus RTK | Sequoia | 448 | 80/80 | 13.5 | |
| 3 | 7 | 10.7.18 | eBee plus RTK | S.O.D.A | 546 | 85/85 | 4.9 |
| 8 | 10.7.18 | eBee plus RTK | Sequoia | 832 | 80/80 | 14.5 | |
| 4 | 9 | 10.7.18 | eBee plus RTK | S.O.D.A | 444 | 85/85 | 4.9 |
| 10 | 10.7.18 | eBee plus RTK | Sequoia | 770 | 80/80 | 14 | |
| 5 | 11 | 28.6.18 | eBee plus RTK | S.O.D.A | 500 | 85/85 | 4.9 |
| 12 | 11.7.18 | eBee plus RTK | S.O.D.A | 765 | 85/85 | 4.9 | |
| 13 | 11.7.18 | eBee plus RTK | S.O.D.A | 875 | 85/85 | 4.9 | |
| 14 | 11.7.18 | eBee plus RTK | S.O.D.A | 611 | 85/85 | 4.9 | |
| 15 | 12.7.18 | eBee plus RTK | S.O.D.A | 829 | 85/85 | 4.9 | |
| 16 | 12.7.18 | eBee plus RTK | S.O.D.A | 735 | 85/85 | 4.9 | |
| 17 | 11.7.18 | eBee plus RTK | Sequoia | 1063 | 80/80 | 13.7 | |
| 18 | 11.7.18 | eBee plus RTK | Sequoia | 196 | 80/80 | 13.7 | |
| 19 | 12.7.18 | eBee plus RTK | Sequoia | 1067 | 80/80 | 13.7 | |
| 20 | 12.7.18 | eBee plus RTK | Sequoia | 851 | 80/80 | 13.7 | |
| 6 | 21 | 09.7.18 | Phantom 4 RTK | Phantom | 199 | 80/80 | 4.0 |
| 22 | 10.7.18 | Matrice 210 | MicaSense | 338 | 88/75 | 9.2 | |
| 7 | 23 | 10.7.18 | Phantom 4 RTK | Phantom | 517 | 80/80 | 3.9 |
| 24 | 10.7.18 | Matrice 210 | MicaSense | 359 | 88/75 | 8.6 | |
| 8 | 25 | 10.7.18 | Phantom 4 RTK | Phantom | 322 | 80/80 | 4.2 |
| 26 | 10.7.18 | Matrice 210 | MicaSense | 309 | 88/75 | 8.8 | |
| 9 | 27 | 10.7.18 | Phantom 4 RTK | Phantom | 320 | 80/80 | 4.3 |
| 28 | 10.7.18 | Matrice 210 | MicaSense | 185 | 88/75 | 8.9 |
| Feature | Source | Description |
|---|---|---|
| Rmean | RGB point cloud | Mean value of the red band |
| Gmean | RGB point cloud | Mean value of the green band |
| Bmean | RGB point cloud | Mean value of the blue band |
| Rsd | RGB point cloud | Standard deviation of the red band |
| Gsd | RGB point cloud | Standard deviation of the green band |
| Bsd | RGB point cloud | Standard deviation of the blue band |
| Red_min | MSP orthomosaic | Minimum value of the red band |
| Red_max | MSP orthomosaic | Maximum value of the red band |
| Red_mean | MSP orthomosaic | Mean value of the red band |
| Red_25 | MSP orthomosaic | 25% percentile of the red band |
| Red_50 | MSP orthomosaic | 50% percentile of the red band |
| Red_75 | MSP orthomosaic | 75% percentile of the red band |
| Green_min | MSP orthomosaic | Minimum value of the green band |
| Green_max | MSP orthomosaic | Maximum value of the green band |
| Green_mean | MSP orthomosaic | Mean value of the green band |
| Green_25 | MSP orthomosaic | 25% percentile of the green band |
| Green_50 | MSP orthomosaic | 50% percentile of the green band |
| Green_75 | MSP orthomosaic | 75% percentile of the green band |
| NIR_min | MSP orthomosaic | Minimum value of the near infrared band |
| NIR_max | MSP orthomosaic | Maximum value of the near infrared band |
| NIR_mean | MSP orthomosaic | Mean value of the near infrared band |
| NIR_25 | MSP orthomosaic | 25% percentile of the near infrared band |
| NIR_50 | MSP orthomosaic | 50% percentile of the near infrared band |
| NIR_75 | MSP orthomosaic | 75% percentile of the near infrared band |
| RE_min | MSP orthomosaic | Minimum value of the red edge band |
| RE_max | MSP orthomosaic | Maximum value of the red edge band |
| RE_mean | MSP orthomosaic | Mean value of the red edge band |
| RE_25 | MSP orthomosaic | 25% percentile of the red edge band |
| RE_50 | MSP orthomosaic | 50% percentile of the red edge band |
| RE_75 | MSP orthomosaic | 75% percentile of the red edge band |
| Blue_min * | MSP orthomosaic | Minimum value of the blue band |
| Blue_max * | MSP orthomosaic | Maximum value of the blue band |
| Blue_mean * | MSP orthomosaic | Mean value of the blue band |
| Blue_25 * | MSP orthomosaic | 25% percentile of the blue band |
| Blue_50 * | MSP orthomosaic | 50% percentile of the blue band |
| Blue_75 * | MSP orthomosaic | 75% percentile of the blue band |
References
- Tikkanen, O.P.; Martikainen, P.; Hyvärinen, E.; Junninen, K.; Kouki, J. Red-listed boreal forest species of Finland: Associations with forest structure, tree species, and decaying wood. Ann. Zool. Fenn. 2006, 43, 373–383. Available online: https://www.jstor.org/stable/23736858 (accessed on 20 March 2021).
- Felton, A.; Lindbladh, M.; Brunet, J.; Fritz, Ö. Replacing coniferous monocultures with mixed-species production stands: An assessment of the potential benefits for forest biodiversity in northern Europe. For. Ecol. Manag. 2010, 260, 939–947. [Google Scholar] [CrossRef]
- Gamfeldt, L.; Snall, T.; Bagchi, R.; Jonsson, M.; Gustafsson, L.; Kjellander, P.; Ruiz-Jaen, M.C.; Fröberg, M.; Stendahl, J.; Philipson, C.D.; et al. Higher levels of multiple ecosystem services are found in forests with more tree species. Nat. Commun. 2013, 4, 1340. [Google Scholar] [CrossRef] [PubMed]
- Brockerhoff, E.G.; Barbaro, L.; Castagneyrol, B.; Forrester, D.I.; Gardiner, B.; González-Olabarria, J.R.; Lyver, P.O.; Meurisse, N.; Oxbrough, A.; Taki, H.; et al. Forest biodiversity, ecosystem functioning and the provision of ecosystem services. Biodiv. Conserv. 2017, 26, 3005–3035. [Google Scholar] [CrossRef] [Green Version]
- Esseen, P.A.; Ehnström, B.; Ericson, L.; Sjöberg, K. Boreal forests. Ecol. Bull. 1997, 46, 16–47. Available online: https://www.jstor.org/stable/20113207 (accessed on 21 March 2021).
- Nilsson, S.G.; Hedin, J.; Niklasson, M. Biodiversity and its assessment in boreal and nemoral forests. Scand. J. For. Res. 2001, 16, 10–26. [Google Scholar] [CrossRef] [Green Version]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Pettorelli, N.; Wegmann, M.; Skidmore, A.; Mücher, S.; Dawson, T.P.; Fernandez, M.; Lucas, R.; Schaepman, M.E.; Wang, T.; O’Connor, B.; et al. Framing the concept of satellite remote sensing essential biodiversity variables: Challenges and future directions. Remote Sens. Ecol. Conserv. 2016, 2, 122–131. [Google Scholar] [CrossRef]
- Rogers, P.C.; Pinno, B.D.; Šebesta, J.; Albrectsen, B.R.; Li, G.; Ivanova, N.; Kusbach, A.; Kuuluvainen, T.; Landhäusser, S.M.; Liu, H.; et al. A global view of aspen: Conservation science for widespread keystone systems. Glob. Ecol. Conserv. 2020, 21, e00828. [Google Scholar] [CrossRef]
- Kivinen, S.; Koivisto, E.; Keski-Saari, S.; Poikolainen, L.; Tanhuanpää, T.; Kuzmin, A.; Viinikka, A.; Heikkinen, R.K.; Pykälä, J.; Virkkala, R.; et al. A keystone species, European aspen (Populus tremula L.), in boreal forests: Ecological role, knowledge needs and mapping using remote sensing. For. Ecol. Manag. 2020, 462, 118008. [Google Scholar] [CrossRef]
- Martikainen, P. Conservation of threatened saproxylic beetles: Significance of retained aspen Populus tremula on clearcut areas. Ecol. Bull. 2001, 49, 205–218. Available online: http://www.jstor.org/stable/20113277 (accessed on 20 March 2021).
- Ranius, T.; Martikainen, P.; Kouki, J. Colonisation of ephemeral forest habitats by specialised species: Beetles and bugs associated with recently dead aspen wood. Biodiv. Conserv. 2011, 20, 2903–2915. [Google Scholar] [CrossRef] [Green Version]
- Kuusinen, M. Epiphytic lichen flora and diversity on Populus tremula in old-growth and managed forests of southern and middle boreal Finland. Annal. Bot. Fenn. 1994, 31, 245–260. Available online: https://www.jstor.org/stable/43922219 (accessed on 20 March 2021).
- Hedenås, H.; Hedström, P. Conservation of epiphytic lichens: Significance of remnant aspen (Populus tremula) trees in clear-cuts. Biol. Conserv. 2007, 135, 388–395. [Google Scholar] [CrossRef]
- Junninen, K.; Penttilä, R.; Martikainen, P. Fallen retention aspen trees on clear-cuts can be important habitats for red-listed polypores: A case study in Finland. Biodiv. Conserv. 2007, 16, 475–490. [Google Scholar] [CrossRef]
- Hanski, I.K. Home ranges and habitat use in the declining flying squirrel Pteromys volans in managed forests. Wildl. Biol. 1998, 4, 33–46. [Google Scholar] [CrossRef]
- Angelstam, P.; Mikusiński, G. Woodpecker assemblages in natural and managed boreal and hemiboreal forest—A review. Annal. Zool. Fenn. 1994, 31, 157–172. Available online: https://www.jstor.org/stable/23735508 (accessed on 20 March 2021).
- Baroni, D.; Korpimäki, E.; Selonen, V.; Laaksonen, T. Tree cavity abundance and beyond: Nesting and food storing sites of the pygmy owl in managed boreal forests. For. Ecol. Manag. 2020, 460, 117818. [Google Scholar] [CrossRef]
- Jonsell, M.; Weslien, J.; Ehnström, B. Substrate requirements of red-listed saproxylic invertebrates in Sweden. Biodivers. Conserv. 1998, 7, 749–764. [Google Scholar] [CrossRef]
- Hyvärinen, E.; Juslén, A.; Kemppainen, E.; Uddström, A.; Liukko, U.-M. Suomen Lajien Uhanalaisuus Punainen Kirja 2019 (The 2019 Red List of Finnish Species); Ympäristöministeriö (Ministry of the Environment) & Suomen Ympäristökeskus (Finnish Environment Institute): Helsinki, Finland, 2019.
- ArtDatabanken. Rodlistade Arter i Sverige 2015 (The 2015 Red List of Swedish Species); ArtDatabanken SLU (Swedish Species Information Centre): Uppsala, Sweden, 2015. [Google Scholar]
- Henriksen, S.; Hilmo, O. Norsk Rødliste for Arter 2015 (The 2015 Norwegian Red List for Species); Artsdatabanken (Norwegian Biodiversity Information Centre): Trondheim, Norway, 2015. [Google Scholar]
- Kouki, J.; Arnold, K.; Martikainen, P. Long-term persistence of aspen—A key host for many threatened species—is endangered in old-growth conservation areas in Finland. J. Nat. Cons. 2004, 12, 41–52. [Google Scholar] [CrossRef]
- Latva-Karjanmaa, T.; Penttilä, R.; Siitonen, J. The demographic structure of European aspen (Populus tremula) populations in managed and old-growth boreal forests in eastern Finland. Can. J. For. Res. 2007, 37, 1070–1081. [Google Scholar] [CrossRef]
- Maltamo, M.; Pesonen, A.; Korhonen, L.; Kouki, J.; Vehmas, M.; Eerikäinen, K. Inventory of aspen trees in spruce dominated stands in conservation area. For. Ecos. 2015, 2, 12. [Google Scholar] [CrossRef] [Green Version]
- Hardenbol, A.; Junninen, K.; Kouki, J. A key tree species for forest biodiversity, European aspen (Populus tremula), is rapidly declining in boreal old-growth forest reserves. For. Ecol. Manag. 2020, 462, 118009. [Google Scholar] [CrossRef]
- Packalén, P.; Temesgen, H.; Maltamo, M. Variable selection strategies for nearest neighbor imputation methods used in remote sensing based forest inventory. Can. J. Rem. Sens. 2007, 38, 557–569. [Google Scholar] [CrossRef]
- Holmgren, J.; Persson, Å.; Söderman, U. Species identification of individual trees by combining high resolution LiDAR data with multi-spectral images. Int. J. Rem. Sens. 2008, 29, 1537–1552. [Google Scholar] [CrossRef]
- Ørka, H.O.; Næsset, E.; Bollandsås, O.M. Effects of different sensors and leaf-on and leaf-off canopy conditions on echo distributions and individual tree properties derived from airborne laser scanning. Remote Sens. Environ. 2010, 114, 1445–1461. [Google Scholar] [CrossRef]
- Trier, Ø.D.; Salberg, A.B.; Kermit, M.; Rudjord, Ø.; Gobakken, T.; Næsset, E.; Aarsten, D. Tree species classification in Norway from airborne hyperspectral and airborne laser scanning data. Eur. J. Remote Sens. 2018, 51, 336–351. [Google Scholar] [CrossRef]
- Viinikka, A.; Hurskainen, P.; Keski-Saari, S.; Kivinen, S.; Tanhuanpää, T.; Mäyrä, J.; Poikolainen, L.; Vihervaara, P.; Kumpula, T. Detecting European aspen (Populus tremula L.) in boreal forests using airborne hyperspectral and airborne laser scanning data. Remote Sens. 2020, 2, 2610. [Google Scholar] [CrossRef]
- Korpela, I.; Ørka, H.O.; Maltamo, M.; Tokola, T.; Hyyppä, J. Tree species classification using airborne LiDAR—effects of stand and tree parameters, downsizing of training set, intensity normalization, and sensor type. Silva Fenn. 2010, 44, 319–339. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Koch, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Fricker, G.A.; Ventura, J.D.; Wolf, J.A.; North, M.P.; Davis, F.W.; Franklin, J. A convolutional neural network classifier identifies tree species in mixed-conifer forest from hyperspectral imagery. Remote Sens. 2019, 11, 2326. [Google Scholar] [CrossRef] [Green Version]
- Grabska, E.; Hostert, P.; Pflugmacher, D.; Ostapowicz, K. Forest stand species mapping using the Sentinel-2 time series. Remote Sens. 2019, 11, 1197. [Google Scholar] [CrossRef] [Green Version]
- Hościło, A.; Lewandowska, A. Mapping forest type and tree species on a regional scale using multi-temporal Sentinel-2 data. Remote Sens. 2019, 11, 929. [Google Scholar] [CrossRef] [Green Version]
- Mäyrä, J.; Keski-Saari, S.; Kivinen, S.; Tanhuanpää, T.; Hurskainen, P.; Kullberg, P.; Poikolainen, L.; Viinikka, A.; Tuominen, S.; Kumpula, T.; et al. Tree species classification from airborne hyperspectral and LiDAR data using 3D convolutional neural networks. Remote Sens. Environ. 2021, 256, 112322. [Google Scholar] [CrossRef]
- Colomina, I.; Molina, P. Unmanned aerial systems for photogrammetry and remote sensing: A review. ISPRS J. Photogr. Remote Sens. 2014, 92, 79–97. [Google Scholar] [CrossRef] [Green Version]
- Tang, L.; Shao, G. Drone remote sensing for forestry research and practices. J. For. Res. 2015, 26, 791–797. [Google Scholar] [CrossRef]
- Torresan, C.; Berton, A.; Carotenuto, F.; Di Gennaro, S.F.; Gioli, B.; Matese, A.; Miglietta, F.; Vagnoli, C.; Zaldei, A.; Wallace, L. Forestry applications of UAVs in Europe: A review. Int. J. Remote Sens. 2017, 38, 2427–2447. [Google Scholar] [CrossRef]
- Sankey, T.; Donager, J.; McVay, J.; Sankey, J.B. UAV lidar and hyperspectral fusion for forest monitoring in the southwestern USA. Remote Sens. Environ. 2017, 195, 30–43. [Google Scholar] [CrossRef]
- Guimarães, N.; Pádua, L.; Marques, P.; Silva, N.; Peres, E.; Sousa, J.J. Forestry remote sensing from Unmanned Aerial Vehicles: A review focusing on the data, processing and potentialities. Remote Sens. 2020, 12, 1046. [Google Scholar] [CrossRef] [Green Version]
- Zarco-Tejada, P.J.; Diaz-Varela, R.; Angileri, V.; Loudjani, P. Tree height quantification using very high resolution imagery acquired from an unmanned aerial vehicle (UAV) and automatic 3D photo-reconstruction methods. Eur. J. Agron. 2014, 55, 89–99. [Google Scholar] [CrossRef]
- Puliti, S.; Ørka, H.O.; Gobakken, T.; Næsset, E. Inventory of small forest areas using Unmanned Aerial System. Remote Sens. 2015, 7, 9632–9654. [Google Scholar] [CrossRef] [Green Version]
- Wallace, L.; Lucieer, A.; Malenovský, Z.; Turner, D.; Vopěnka, P. Assessment of forest structure using two UAV techniques: A comparison of airborne laser scanning and structure from motion (SfM) point clouds. Forests 2016, 7, 62. [Google Scholar] [CrossRef] [Green Version]
- Goodbody, T.R.H.; Coops, N.C.; Marshall, P.L.; Tompalski, P.; Crawford, P. Unmanned aerial systems for precision forest inventory purposes: A review and case study. For. Chron. 2017, 93, 71–81. [Google Scholar] [CrossRef] [Green Version]
- Jayathunga, S.; Owari, T.; Tsuyuki, S.; Hirata, Y. Potential of UAV photogrammetry for characterization of forest canopy structure in uneven-aged mixed conifer—Broadleaf forests. Int. J. Remote Sens. 2020, 41, 53–73. [Google Scholar] [CrossRef]
- Miyoshi, G.T.; Arruda, M.d.S.; Osco, L.P.; Junior, J.M.; Gonçalves, D.N.; Imai, N.N.; Tommaselli, A.M.G.; Honkavaara, E.; Gonçalves, W.N. A Novel Deep Learning Method to Identify Single Tree Species in UAV-Based Hyperspectral Images. Remote Sens. 2020, 12, 1294. [Google Scholar] [CrossRef] [Green Version]
- Getzin, S.; Wiegand, K.; Schöning, I. Assessing biodiversity in forests using very high-resolution images and unmanned aerial vehicles. Meth. Ecol. Evol. 2012, 3, 397–404. [Google Scholar] [CrossRef]
- Anderson, K.; Gaston, K.J. Lightweight unmanned aerial vehicles will revolutionize spatial ecology. Front. Ecol. Environ. 2013, 11, 138–146. [Google Scholar] [CrossRef] [Green Version]
- López, J.J.; Mulero-Pázmány, M. Drones for conservation in protected areas: Present and future. Drones 2019, 3, 10. [Google Scholar] [CrossRef] [Green Version]
- Baena, S.; Moat, J.; Whaley, O.; Boyd, D.S. Identifying species from the air: UAVs and the very high resolution challenge for plant conservation. PLoS ONE 2017, 12, e0188714. [Google Scholar] [CrossRef] [Green Version]
- Michez, A.; Piégay, H.; Lisein, J.; Claessens, H.; Lejeune, P. Classification of riparian forest species and health condition using multi-temporal and hyperspatial imagery from unmanned aerial system. Environ. Mon. Assess. 2016, 188, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gini, R.; Passoni, D.; Pinto, L.; Sona, G. Use of unmanned aerial systems for multispectral survey and tree classification: A test in a park area of northern Italy. Eur. J. Remote Sens. 2014, 47, 251–269. [Google Scholar] [CrossRef]
- Bagaram, M.B.; Giuliarelli, D.; Chirici, G.; Giannetti, F.; Barbati, A. UAV Remote Sensing for Biodiversity Monitoring: Are Forest Canopy Gaps Good Covariates? Remote Sens. 2018, 10, 9139. [Google Scholar] [CrossRef]
- Briechle, S.; Krzystek, P.; Vosselman, G. Classification of tree species and standing dead trees by fusing UAV-based lidar data and multispectral imagery in the 3D deep neural network PointNet++. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2020, 5, 203–210. [Google Scholar] [CrossRef]
- Thiel, C.; Mueller, M.M.; Epple, L.; Thau, C.; Hese, S.; Voltersen, M.; Henkel, A. UAS Imagery-Based Mapping of Coarse Wood Debris in a Natural Deciduous Forest in Central Germany (Hainich National Park). Remote Sens. 2020, 12, 3293. [Google Scholar] [CrossRef]
- Nevalainen, O.; Honkavaara, E.; Tuominen, S.; Viljanen, N.; Hakala, T.; Yu, X.; Hyyppä, J.; Saari, H.; Pölönen, I.; Imai, N.N.; et al. Individual Tree Detection and Classification with UAV-Based Photogrammetric Point Clouds and Hyperspectral Imaging. Remote Sens. 2017, 9, 185. [Google Scholar] [CrossRef] [Green Version]
- Nezami, S.; Khoramshahi, E.; Nevalainen, O.; Pölönen, I.; Honkavaara, E. Tree Species Classification of Drone Hyperspectral and RGB Imagery with Deep Learning Convolutional Neural Networks. Remote. Sens. 2020, 12, 1070. [Google Scholar] [CrossRef] [Green Version]
- Tuominen, S.; Näsi, R.; Honkavaara, E.; Balazs, A.; Hakala, T.; Viljanen, N.; Pölönen, I.; Saari, H.; Ojanen, H. Assessment of Classifiers and Remote Sensing Features of Hyperspectral Imagery and Stereo-Photogrammetric Point Clouds for Recognition of Tree Species in a Forest Area of High Species Diversity. Remote Sens. 2018, 10, 714. [Google Scholar] [CrossRef] [Green Version]
- Saarinen, N.; Vastaranta, M.; Näsi, R.; Rosnell, T.; Hakala, T.; Honkavaara, E.; Wulder, M.A.; Luoma, V.; Tommaselli, A.M.G.; Imai, N.N.; et al. Assessing Biodiversity in Boreal Forests with UAV-Based Photogrammetric Point Clouds and Hyperspectral Imaging. Remote Sens. 2018, 10, 338. [Google Scholar] [CrossRef] [Green Version]
- Gini, R.; Sona, G.; Ronchetti, G.; Passoni, D.; Pinto, L. Improving tree species classification using UAS multispectral images and texture measures. ISPRS Int. J. Geo-Inf. 2018, 7, 315. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Shen, X.; Cao, L.; Coops, N.C.; Goodbody, T.R.H.; Zhong, T.; Zhao, W.; Sun, Q.; Ba, S.; Zhang, Z.; et al. Tree species classification using UAS-based digital aerial photogrammetry point clouds and multispectral imageries in subtropical natural forests. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102173. [Google Scholar] [CrossRef]
- Franklin, S.E.; Ahmed, O.S. Deciduous tree species classification using object-based analysis and machine learning with unmanned aerial vehicle multispectral data. Int. J. Remote Sens. 2018, 39, 5236–5245. [Google Scholar] [CrossRef]
- Brown, D.C. Close-range camera calibration. Photogramm. Eng. 1971, 37, 855–866. [Google Scholar]
- Puliti, S.; Gobakken, T.; Ørka, H.O.; Næsset, E. Assessing 3D point clouds from aerial photographs for species-specific forest inventories. Scand. J. For. Res. 2017, 32, 68–79. [Google Scholar] [CrossRef]
- Isenburg, M. “LAStools-Efficient LiDAR Processing Software” (Version 200509, Academic License). 2021. Available online: https://rapidlasso.com/LAStools (accessed on 20 March 2021).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Silva, C.A.; Crookston, N.L.; Hudak, A.T.; Vierling, L.A.; Klauberg, C.; Cardil, A. rLiDAR: LiDAR Data Processing and Visualization. Available online: https://cran.r-project.org/package=rLiDAR (accessed on 20 March 2021).
- Yu, B.; Ostland, M.; Gong, P.; Pu, R. Penalized discriminant analysis of in situ hyperspectral data for conifer species recognition. IEEE Transact. Geosci. Remote Sens. 1999, 37, 2569–2577. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, M. Caret: Classification and Regression Training. Misc Functions for Training and Plotting Classification and Regression Models. Available online: http://CRAN.R-project.org/package=caret (accessed on 20 March 2021).
- Mountrakis, G.; Im, J.; Ogole, C. Support vector machines in remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Kuhn, M. The Caret Package Documentation, 2019-03-27. Available online: http://topepo.github.io/caret/index.html (accessed on 4 March 2021).
- Molnar, C. Interpretable Machine Learning. A Guide for Making Black Box Models Explainable 2019. Available online: https://christophm.github.io/interpretable-ml-book/ (accessed on 29 January 2021).
- Fisher, A.; Rudin, C.; Dominici, F. All models are wrong, but many are useful: Learning a variable’s importance by studying an entire class of prediction models simultaneously. J. Mach. Learn. Res. 2019, 20, 1–81. Available online: http://www.jmlr.org/papers/v20/18-760.html (accessed on 21 March 2021).
- Xu, R.; Li, C.; Paterson, A.H. Multispectral imaging and unmanned aerial systems for cotton plant phenotyping. PLoS ONE 2019, 14, e0205083. [Google Scholar] [CrossRef] [Green Version]
- Roth, L.; Streit, B. Predicting cover crop biomass by lightweight uas-based rgb and nir photography: An applied photogrammetric approach. Precis. Agric. 2018, 19, 93–114. [Google Scholar] [CrossRef] [Green Version]
- Ashapure, A.; Jung, J.; Chang, A.; Oh, S.; Maeda, M.; Landivar, J. A Comparative Study of RGB and Multispectral Sensor-Based Cotton Canopy Cover Modelling Using Multi-Temporal UAS Data. Remote Sens. 2019, 11, 2757. [Google Scholar] [CrossRef] [Green Version]
- Zhen, Z.; Quackenbush, L.J.; Zhang, L. Trends in automatic individual tree crown detection and delineation-evolution of LiDAR data. Remote Sens. 2016, 8, 333. [Google Scholar] [CrossRef] [Green Version]
- Dalponte, M.; Ørka, H.O.; Ene, L.T.; Gobakken, T.; Næsset, E. Tree Crown Delineation and Tree Species Classification in Boreal Forests Using Hyperspectral and ALS Data. Remote Sens. Environ. 2014, 140, 306–317. [Google Scholar] [CrossRef]
- Kwak, D.A.; Lee, W.K.; Lee, J.H.; Biging, G.S.; Gong, P. Detection of individual trees and estimation of tree height using LiDAR data. J. Forest Res. 2007, 12, 425–434. [Google Scholar] [CrossRef]
- Tanhuanpää, T.; Yu, X.; Luoma, V.; Saarinen, N.; Raisio, J.; Hyyppä, J.; Kumpula, T.; Holopainen, M. Effect of canopy structure on the performance of tree mapping methods in urban parks. Urban For. Urban Green. 2019, 44, 126441. [Google Scholar] [CrossRef]
- Lisein, J.; Michez, A.; Claessens, H.; Lejeune, P. Discrimination of deciduous tree species from time series of unmanned aerial system imagery. PLoS ONE 2015, 10, e0141006. [Google Scholar] [CrossRef] [PubMed]
- Weil, G.; Lensky, I.; Resheff, Y.; Levin, N. Optimizing the Timing of Unmanned Aerial Vehicle Image Acquisition for Applied Mapping of Woody Vegetation Species Using Feature Selection. Remote Sens. 2017, 9, 1130. [Google Scholar] [CrossRef] [Green Version]
- Heide, O.M. Daylength and thermal time responses of budburst during dormancy release in some northern deciduous trees. Physiol. Plant. 1993, 88, 531–540. [Google Scholar] [CrossRef]
- Brown, L.A.; Dash, J.; Ogutu, B.O.; Richardson, A.D. On the relationship between continuous measures of canopy greenness derived using near-surface remote sensing and satellite-derived vegetation products. Agric. For. Meteorol. 2017, 247, 280–292. [Google Scholar] [CrossRef] [Green Version]
- Fawcett, D.; Bennie, J.; Anderson, K. Monitoring spring phenology of individual tree crowns using drone—Acquired NDVI data. Remote Sens. Ecol. Conserv. 2020. [Google Scholar] [CrossRef]
- Gu, W.D.; Kuusinen, M.; Konttinen, T.; Hanski, I. Spatial pattern in the occurrence of the lichen Lobaria pulmonaria in managed and virgin boreal forests. Ecography 2001, 24, 139–150. [Google Scholar] [CrossRef]
- Suominen, O.; Edenius, L.; Ericsson, G.; de Dios, V.R. Gastropod diversity in aspen stands in coastal northern Sweden. For. Ecol. Manag. 2003, 175, 403–412. [Google Scholar] [CrossRef]
- Kattenborn, T.; Lopatin, J.; Förster, M.; Braun, A.C.; Fassnacht, F.E. UAV data as alternative to field sampling to map woody invasive species based on combined Sentinel-1 and Sentinel-2 data. Remote Sens. Environ. 2019, 227, 61–73. [Google Scholar] [CrossRef]
- Riihimäki, H.; Luoto, M.; Heiskanen, J. Estimating Fractional Cover of Tundra Vegetation at Multiple Scales Using Unmanned Aerial Systems and Optical Satellite Data. Remote Sens. Environ. 2019, 224, 119–132. [Google Scholar] [CrossRef]
- Gränzig, T.; Fassnacht, F.E.; Kleinschmit, B.; Förster, M. Mapping the fractional coverage of the invasive shrub Ulex europaeus with multi-temporal Sentinel-2 imagery utilizing UAV orthoimages and a new spatial optimization approach. Int. J. Appl. Earth Obs. Geoinf. 2021, 96, 102281. [Google Scholar] [CrossRef]





| Species | MicaSense Dataset | Sequoia Dataset |
|---|---|---|
| Scots pine (Pinus sylvestris L.) | 75 | 120 |
| Norway spruce (Picea abies L.) | 66 | 120 |
| Downy birch (Betula pubescens L.) and Silver birch (Betula pendula) | 75 | 107 |
| Aspen (Populus tremula L.) | 90 | 118 |
| Total: | 306 | 465 |
| Camera (Feature Set) | Number of Features | F1-Score (%) | Overall Accuracy (%) | Kappa | |
|---|---|---|---|---|---|
| Aspen | Other | ||||
| MicaSense (RGB) | 6 | 84 | 93 | 90.1 | 0.77 |
| MicaSense (MSP) | 30 | 81 | 91 | 87.9 | 0.72 |
| MicaSense (RGB+MSP) | 32 | 82 | 93 | 90.1 | 0.76 |
| Sequoia (RGB) | 6 | 79 | 93 | 89.9 | 0.73 |
| Sequoia (MSP) | 21 | 68 | 90 | 84.2 | 0.57 |
| Sequoia (RGB+MSP) | 29 | 77 | 93 | 89.2 | 0.70 |
| (a) | |||||
| MicaSense (RGB) | Predicted | ||||
| Aspen | Other | Total | UA (%) | ||
| Observed | Aspen | 23 | 5 | 28 | 82.1 |
| Other | 4 | 59 | 63 | 93.7 | |
| Total | 27 | 64 | 91 | ||
| PA (%) | 85.2 | 92.2 | |||
| OA (%) | 90.1 | ||||
| (b) | |||||
| Sequoia (RGB) | Predicted | ||||
| Aspen | Other | Total | UA (%) | ||
| Observed | Aspen | 27 | 6 | 33 | 81.8 |
| Other | 8 | 98 | 106 | 92.5 | |
| Total | 35 | 104 | 139 | ||
| PA (%) | 77.1 | 94.2 | |||
| OA (%) | 88.9 | ||||
| Camera (Feature Set) | Number of Features | F1-Score (%) | OA (%) | Kappa | |||
|---|---|---|---|---|---|---|---|
| Aspen | Birch | Pine | Spruce | ||||
| MicaSense (RGB) | 6 | 82 | 78 | 79 | 76 | 78.9 | 0.72 |
| MicaSense (MSP) | 24 | 84 | 76 | 71 | 91 | 81.1 | 0.75 |
| MicaSense (RGB+MSP) | 28 | 86 | 78 | 80 | 88 | 83.3 | 0.78 |
| Sequoia (RGB) | 6 | 78 | 64 | 89 | 93 | 81.3 | 0.75 |
| Sequoia (MSP) | 17 | 63 | 57 | 72 | 79 | 68.3 | 0.58 |
| Sequoia (RGB+MSP) | 26 | 80 | 69 | 81 | 94 | 81.3 | 0.75 |
| (a) | |||||||
| MicaSense (RGB+MSP) | Predicted | UA (%) | |||||
| Aspen | Birch | Pine | Spruce | Total | |||
| Observed | Aspen | 24 | 3 | 1 | 1 | 29 | 82.8 |
| Birch | 1 | 16 | 1 | 1 | 23 | 84.2 | |
| Pine | 1 | 3 | 16 | 1 | 16 | 76.2 | |
| Spruce | 1 | 0 | 1 | 19 | 22 | 90.5 | |
| Total | 27 | 22 | 19 | 22 | 90 | ||
| PA (%) | 88.9 | 72.7 | 84.2 | 86.4 | |||
| OA (%) | 83.3 | ||||||
| (b) | |||||||
| Sequoia (RGB+MSP) | Predicted | UA (%) | |||||
| Aspen | Birch | Pine | Spruce | Total | |||
| Observed | Aspen | 28 | 6 | 1 | 0 | 35 | 80.0 |
| Birch | 4 | 22 | 5 | 1 | 32 | 68.8 | |
| Pine | 3 | 3 | 30 | 2 | 38 | 78.9 | |
| Spruce | 0 | 1 | 0 | 33 | 34 | 97.1 | |
| Total | 35 | 32 | 36 | 36 | 139 | ||
| PA (%) | 80.0 | 68.8 | 83.3 | 91.7 | |||
| OA (%) | 81.3 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuzmin, A.; Korhonen, L.; Kivinen, S.; Hurskainen, P.; Korpelainen, P.; Tanhuanpää, T.; Maltamo, M.; Vihervaara, P.; Kumpula, T. Detection of European Aspen (Populus tremula L.) Based on an Unmanned Aerial Vehicle Approach in Boreal Forests. Remote Sens. 2021, 13, 1723. https://doi.org/10.3390/rs13091723
Kuzmin A, Korhonen L, Kivinen S, Hurskainen P, Korpelainen P, Tanhuanpää T, Maltamo M, Vihervaara P, Kumpula T. Detection of European Aspen (Populus tremula L.) Based on an Unmanned Aerial Vehicle Approach in Boreal Forests. Remote Sensing. 2021; 13(9):1723. https://doi.org/10.3390/rs13091723
Chicago/Turabian StyleKuzmin, Anton, Lauri Korhonen, Sonja Kivinen, Pekka Hurskainen, Pasi Korpelainen, Topi Tanhuanpää, Matti Maltamo, Petteri Vihervaara, and Timo Kumpula. 2021. "Detection of European Aspen (Populus tremula L.) Based on an Unmanned Aerial Vehicle Approach in Boreal Forests" Remote Sensing 13, no. 9: 1723. https://doi.org/10.3390/rs13091723