Two-Stage Convolutional Neural Networks for Diagnosing the Severity of Alternaria Leaf Blotch Disease of the Apple Tree
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Annotation and Examination
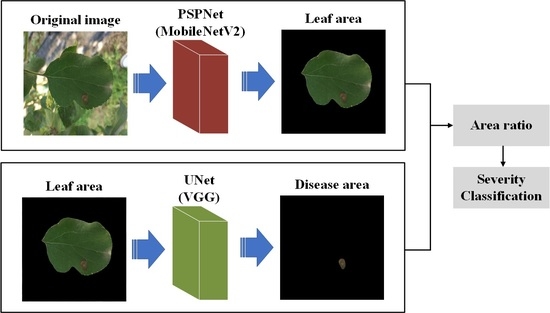
2.2. Deep-Learning Algorithms
2.3. Evaluation Metrics
2.4. Apple Alternaria Leaf Blotch Severity Classification Method
2.5. Equipment
3. Results
3.1. Model Training
3.2. Leaf and Disease-Area Identification
3.3. Examination of Apple Alternaria Leaf Blotch Severity
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bi, C.; Wang, J.; Duan, Y.; Fu, B.; Kang, J.-R.; Shi, Y. MobileNet based apple leaf diseases identification. Mob. Netw. Appl. 2020, 27, 172–180. [Google Scholar] [CrossRef]
- Norelli, J.L.; Farrell, R.E.; Bassett, C.L.; Baldo, A.M.; Lalli, D.A.; Aldwinckle, H.S.; Wisniewski, M.E. Rapid transcriptional response of apple to fire blight disease revealed by cDNA suppression subtractive hybridization analysis. Tree Genet. Genomes 2009, 5, 27–40. [Google Scholar] [CrossRef]
- Hou, Y.; Zhang, X.; Zhang, N.; Naklumpa, W.; Zhao, W.; Liang, X.; Zhang, R.; Sun, G.; Gleason, M. Genera Acremonium and Sarocladium cause brown spot on bagged apple fruit in China. Plant Dis. 2019, 103, 1889–1901. [Google Scholar] [CrossRef] [PubMed]
- Xu, X. Modelling and forecasting epidemics of apple powdery mildew (Podosphaera leucotricha). Plant Pathol. 1999, 48, 462–471. [Google Scholar] [CrossRef]
- Grove, G.G.; Eastwell, K.C.; Jones, A.L.; Sutton, T.B. 18 Diseases of Apple. In Apples: Botany, Production, and Uses; CABI Publishing Oxon: Wallingford, UK, 2003; pp. 468–472. [Google Scholar]
- Ji, M.; Zhang, K.; Wu, Q.; Deng, Z. Multi-label learning for crop leaf diseases recognition and severity estimation based on convolutional neural networks. Soft Comput. 2020, 24, 15327–15340. [Google Scholar] [CrossRef]
- Roberts, J.W. Morphological characters of Alternaria mali Roberts. J. Agric. Res. 1924, 27, 699–708. [Google Scholar]
- Jung, K.-H. Growth inhibition effect of pyroligneous acid on pathogenic fungus, Alternaria mali, the agent of Alternaria blotch of apple. Biotechnol. Bioprocess Eng. 2007, 12, 318–322. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Y.; Liu, H.; Han, J. Isolation of Alternaria mali Roberts and its sensitivities to four fungicides. J. Shanxi Agric. Univ. 2004, 24, 382–384. [Google Scholar]
- Zhang, C.-X.; Tian, Y.; Cong, P.-H. Proteome analysis of pathogen-responsive proteins from apple leaves induced by the alternaria blotch Alternaria alternata. PLoS ONE 2015, 10, e0122233. [Google Scholar] [CrossRef]
- Lu, Y.; Song, S.; Wang, R.; Liu, Z.; Meng, J.; Sweetman, A.J.; Jenkins, A.; Ferrier, R.C.; Li, H.; Luo, W. Impacts of soil and water pollution on food safety and health risks in China. Environ. Int. 2015, 77, 5–15. [Google Scholar] [CrossRef] [Green Version]
- Sutton, T.B. Changing options for the control of deciduous fruit tree diseases. Annu. Rev. Phytopathol. 1996, 34, 527–547. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, G.; Satya, S.; Naik, S. Food processing a tool to pesticide residue dissipation—A review. Food Res. Int. 2009, 42, 26–40. [Google Scholar] [CrossRef]
- Mahmud, M.S.; Zahid, A.; He, L.; Martin, P. Opportunities and possibilities of developing an advanced precision spraying system for tree fruits. Sensors 2021, 21, 3262. [Google Scholar] [CrossRef] [PubMed]
- Pathania, A.; Rialch, N.; Sharma, P. Marker-assisted selection in disease resistance breeding: A boon to enhance agriculture production. In Current Developments in Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2017; pp. 187–213. [Google Scholar]
- Jiang, P.; Chen, Y.; Liu, B.; He, D.; Liang, C. Real-time detection of apple leaf diseases using deep learning approach based on improved convolutional neural networks. IEEE Access 2019, 7, 59069–59080. [Google Scholar] [CrossRef]
- Dutot, M.; Nelson, L.; Tyson, R. Predicting the spread of postharvest disease in stored fruit, with application to apples. Postharvest Biol. Technol. 2013, 85, 45–56. [Google Scholar] [CrossRef]
- Bock, C.; Poole, G.; Parker, P.; Gottwald, T. Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. Crit. Rev. Plant Sci. 2010, 29, 59–107. [Google Scholar] [CrossRef]
- Ampatzidis, Y.; De Bellis, L.; Luvisi, A. iPathology: Robotic applications and management of plants and plant diseases. Sustainability 2017, 9, 1010. [Google Scholar] [CrossRef] [Green Version]
- Su, W.-H.; Yang, C.; Dong, Y.; Johnson, R.; Page, R.; Szinyei, T.; Hirsch, C.D.; Steffenson, B.J. Hyperspectral imaging and improved feature variable selection for automated determination of deoxynivalenol in various genetic lines of barley kernels for resistance screening. Food Chem. 2021, 343, 128507. [Google Scholar] [CrossRef]
- Su, W.-H.; Sheng, J.; Huang, Q.-Y. Development of a Three-Dimensional Plant Localization Technique for Automatic Differentiation of Soybean from Intra-Row Weeds. Agriculture 2022, 12, 195. [Google Scholar] [CrossRef]
- Su, W.-H. Advanced Machine Learning in Point Spectroscopy, RGB-and hyperspectral-imaging for automatic discriminations of crops and weeds: A review. Smart Cities 2020, 3, 767–792. [Google Scholar] [CrossRef]
- Su, W.-H.; Slaughter, D.C.; Fennimore, S.A. Non-destructive evaluation of photostability of crop signaling compounds and dose effects on celery vigor for precision plant identification using computer vision. Comput. Electron. Agric. 2020, 168, 105155. [Google Scholar] [CrossRef]
- Su, W.-H.; Fennimore, S.A.; Slaughter, D.C. Development of a systemic crop signalling system for automated real-time plant care in vegetable crops. Biosyst. Eng. 2020, 193, 62–74. [Google Scholar] [CrossRef]
- Su, W.-H.; Sun, D.-W. Advanced analysis of roots and tubers by hyperspectral techniques. In Advances in Food and Nutrition Research; Elsevier: Amsterdam, The Netherlands, 2019; Volume 87, pp. 255–303. [Google Scholar]
- Su, W.-H.; Zhang, J.; Yang, C.; Page, R.; Szinyei, T.; Hirsch, C.D.; Steffenson, B.J. Evaluation of Mask RCNN for Learning to Detect Fusarium Head Blight in Wheat Images. In Proceedings of the 2020 ASABE Annual International Virtual Meeting, Online, 13–15 July 2020; American Society of Agricultural and Biological Engineers: St. Joseph, MI, USA, 2020; p. 1. [Google Scholar]
- Su, W.-H. Crop plant signaling for real-time plant identification in smart farm: A systematic review and new concept in artificial intelligence for automated weed control. Artif. Intell. Agric. 2020, 4, 262–271. [Google Scholar] [CrossRef]
- Gargade, A.; Khandekar, S. Custard apple leaf parameter analysis, leaf diseases, and nutritional deficiencies detection using machine learning. In Advances in Signal and Data Processing; Springer: Berlin/Heidelberg, Germany, 2021; pp. 57–74. [Google Scholar]
- Jan, M.; Ahmad, H. Image Features Based Intelligent Apple Disease Prediction System: Machine Learning Based Apple Disease Prediction System. Int. J. Agric. Environ. Inf. Syst. (IJAEIS) 2020, 11, 31–47. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, Y.; He, D.; Li, Y. Identification of apple leaf diseases based on deep convolutional neural networks. Symmetry 2018, 10, 11. [Google Scholar] [CrossRef] [Green Version]
- Chao, X.; Sun, G.; Zhao, H.; Li, M.; He, D. Identification of apple tree leaf diseases based on deep learning models. Symmetry 2020, 12, 1065. [Google Scholar] [CrossRef]
- Baliyan, A.; Kukreja, V.; Salonki, V.; Kaswan, K.S. Detection of Corn Gray Leaf Spot Severity Levels using Deep Learning Approach. In Proceedings of the 2021 9th International Conference on Reliability, Infocom Technologies and Optimization (Trends and Future Directions) (ICRITO), Noida, India, 3–4 September 2021; pp. 1–5. [Google Scholar]
- Ozguven, M.M.; Adem, K. Automatic detection and classification of leaf spot disease in sugar beet using deep learning algorithms. Phys. A Stat. Mech. Its Appl. 2019, 535, 122537. [Google Scholar] [CrossRef]
- Esgario, J.G.; Krohling, R.A.; Ventura, J.A. Deep learning for classification and severity estimation of coffee leaf biotic stress. Comput. Electron. Agric. 2020, 169, 105162. [Google Scholar] [CrossRef] [Green Version]
- Verma, S.; Chug, A.; Singh, A.P. Application of convolutional neural networks for evaluation of disease severity in tomato plant. J. Discret. Math. Sci. Cryptogr. 2020, 23, 273–282. [Google Scholar] [CrossRef]
- Su, W.-H.; Zhang, J.; Yang, C.; Page, R.; Szinyei, T.; Hirsch, C.D.; Steffenson, B.J. Automatic evaluation of wheat resistance to fusarium head blight using dual mask-RCNN deep learning frameworks in computer vision. Remote Sens. 2020, 13, 26. [Google Scholar] [CrossRef]
- Wang, C.; Du, P.; Wu, H.; Li, J.; Zhao, C.; Zhu, H. A cucumber leaf disease severity classification method based on the fusion of DeepLabV3+ and U-Net. Comput. Electron. Agric. 2021, 189, 106373. [Google Scholar] [CrossRef]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Picon, A.; San-Emeterio, M.G.; Bereciartua-Perez, A.; Klukas, C.; Eggers, T.; Navarra-Mestre, R. Deep learning-based segmentation of multiple species of weeds and corn crop using synthetic and real image datasets. Comput. Electron. Agric. 2022, 194, 106719. [Google Scholar] [CrossRef]
- Lv, Q.; Wang, H. Cotton Boll Growth Status Recognition Method under Complex Background Based on Semantic Segmentation. In Proceedings of the 2021 4th International Conference on Robotics, Control and Automation Engineering (RCAE), Wuhan, China, 4–6 November 2021; pp. 50–54. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Chen, L.-C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Xia, X.; Xu, C.; Nan, B. Inception-v3 for flower classification. In Proceedings of the 2017 2nd International Conference on Image, Vision and Computing (ICIVC), Chengdu, China, 2–4 June 2017; pp. 783–787. [Google Scholar]
- Garcia-Garcia, A.; Orts-Escolano, S.; Oprea, S.; Villena-Martinez, V.; Garcia-Rodriguez, J. A review on deep learning techniques applied to semantic segmentation. arXiv 2017, arXiv:1704.06857. [Google Scholar]
- Hayit, T.; Erbay, H.; Varçın, F.; Hayit, F.; Akci, N. Determination of the severity level of yellow rust disease in wheat by using convolutional neural networks. J. Plant Pathol. 2021, 103, 923–934. [Google Scholar] [CrossRef]
- Prabhakar, M.; Purushothaman, R.; Awasthi, D.P. Deep learning based assessment of disease severity for early blight in tomato crop. Multimed. Tools Appl. 2020, 79, 28773–28784. [Google Scholar] [CrossRef]
- Wu, Z.; Yang, R.; Gao, F.; Wang, W.; Fu, L.; Li, R. Segmentation of abnormal leaves of hydroponic lettuce based on DeepLabV3+ for robotic sorting. Comput. Electron. Agric. 2021, 190, 106443. [Google Scholar] [CrossRef]
- Ji, M.; Wu, Z. Automatic detection and severity analysis of grape black measles disease based on deep learning and fuzzy logic. Comput. Electron. Agric. 2022, 193, 106718. [Google Scholar] [CrossRef]
- Nigam, S.; Jain, R.; Prakash, S.; Marwaha, S.; Arora, A.; Singh, V.K.; Singh, A.K.; Prakasha, T. Wheat Disease Severity Estimation: A Deep Learning Approach. In Proceedings of the International Conference on Internet of Things and Connected Technologies, Patna, India, 29–30 July 2021; pp. 185–193. [Google Scholar]
- Ramcharan, A.; McCloskey, P.; Baranowski, K.; Mbilinyi, N.; Mrisho, L.; Ndalahwa, M.; Legg, J.; Hughes, D.P. A mobile-based deep learning model for cassava disease diagnosis. Front. Plant Sci. 2019, 10, 272. [Google Scholar] [CrossRef] [Green Version]
- Hu, G.; Wang, H.; Zhang, Y.; Wan, M. Detection and severity analysis of tea leaf blight based on deep learning. Comput. Electr. Eng. 2021, 90, 107023. [Google Scholar] [CrossRef]
- Zeng, Q.; Ma, X.; Cheng, B.; Zhou, E.; Pang, W. Gans-based data augmentation for citrus disease severity detection using deep learning. IEEE Access 2020, 8, 172882–172891. [Google Scholar] [CrossRef]









| Modeling Parameters | Values |
|---|---|
| Number of training samples | 4004 |
| Number of validation samples | 494 |
| Number of test samples | 444 |
| Number of overall test samples | 440 |
| Input size | 512 × 512 |
| Training number of epochs | 100 |
| Base learning rate | 0.0001 |
| Image input batch size | 2 |
| Gamma | 0.1 |
| Number of classes | 2 |
| Maximum iterations | 2224 |
| Model | Backbone | Precision | Recall | MIoU |
|---|---|---|---|---|
| DeeplabV3+ | MobileNetV2 | 99.00% | 99.04% | 98.06% |
| Xception | 98.74% | 98.86% | 97.63% | |
| PSPNet | MobileNetV2 | 99.15% | 99.26% | 98.42% |
| ResNet | 99.10% | 99.21% | 98.33% | |
| UNet | ResNet | 99.12% | 99.27% | 98.41% |
| VGG | 99.07% | 99.24% | 98.32% |
| Model | Backbone | Precision | Recall | MIoU |
|---|---|---|---|---|
| DeeplabV3+ | MobileNetV2 | 95.04% | 94.23% | 90.30% |
| Xception | 95.47% | 91.51% | 88.32% | |
| PSPNet | MobileNetV2 | 93.53% | 93.80% | 88.74% |
| ResNet | 93.99% | 93.11% | 88.55% | |
| UNet | ResNet | 95.92% | 94.55% | 91.27% |
| VGG | 95.84% | 95.54% | 92.05% |
| Disease Classification | Correct Grading | Data Quantity | Accuracy |
|---|---|---|---|
| Healthy | 44 | 44 | 100% |
| Early | 179 | 183 | 97.81% |
| Mild | 90 | 98 | 91.54% |
| Moderate | 70 | 73 | 95.89% |
| Severe | 47 | 48 | 97.92% |
| Total | 430 | 446 | 96.41% |
| References | Plant | Model | Disease Levels | Accuracy (%) |
|---|---|---|---|---|
| Hayit et al. [50] | Wheat | Xception | 0, R, MR, MRMS, MS, S | 91 |
| Nigam et al. [54] | Wheat | Proposal modified CNNs | Healthy stage, early stage, middle stage and end-stage | 96.42 |
| Ramcharan et al. [55] | Cassava | MobileNet | Mild symptoms (A–C) and pronounced symptoms | 84.70 |
| Hu et al. [56] | Tea | VGG16 | Mild and severe | 90 |
| Zeng et al. [57] | Citrus | AlexNet, InceptionV3, ResNet | Early, mild, moderate, severe | 92.60 |
| Prabhakar et al. [51] | Tomato | AlexNet, VGGNet, GoogleNet ResNet | Healthy, mild, moderate, severe | 94.60 |
| Ji and Wu [53] | Grape | DeepLabV3+ (ResNet-50) | Healthy, mild, medium, severe | 97.75 |
| Proposed method | Apple | PSPNet (MobileNetV2) and UNet (VGG) | Healthy, early, mild, moderate, severe | 96.41 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.-Y.; Fan, K.-J.; Su, W.-H.; Peng, Y. Two-Stage Convolutional Neural Networks for Diagnosing the Severity of Alternaria Leaf Blotch Disease of the Apple Tree. Remote Sens. 2022, 14, 2519. https://doi.org/10.3390/rs14112519
Liu B-Y, Fan K-J, Su W-H, Peng Y. Two-Stage Convolutional Neural Networks for Diagnosing the Severity of Alternaria Leaf Blotch Disease of the Apple Tree. Remote Sensing. 2022; 14(11):2519. https://doi.org/10.3390/rs14112519
Chicago/Turabian StyleLiu, Bo-Yuan, Ke-Jun Fan, Wen-Hao Su, and Yankun Peng. 2022. "Two-Stage Convolutional Neural Networks for Diagnosing the Severity of Alternaria Leaf Blotch Disease of the Apple Tree" Remote Sensing 14, no. 11: 2519. https://doi.org/10.3390/rs14112519