Ultra-High-Resolution UAV-Based Detection of Alternaria solani Infections in Potato Fields
Abstract
:1. Introduction
2. Materials and Methods
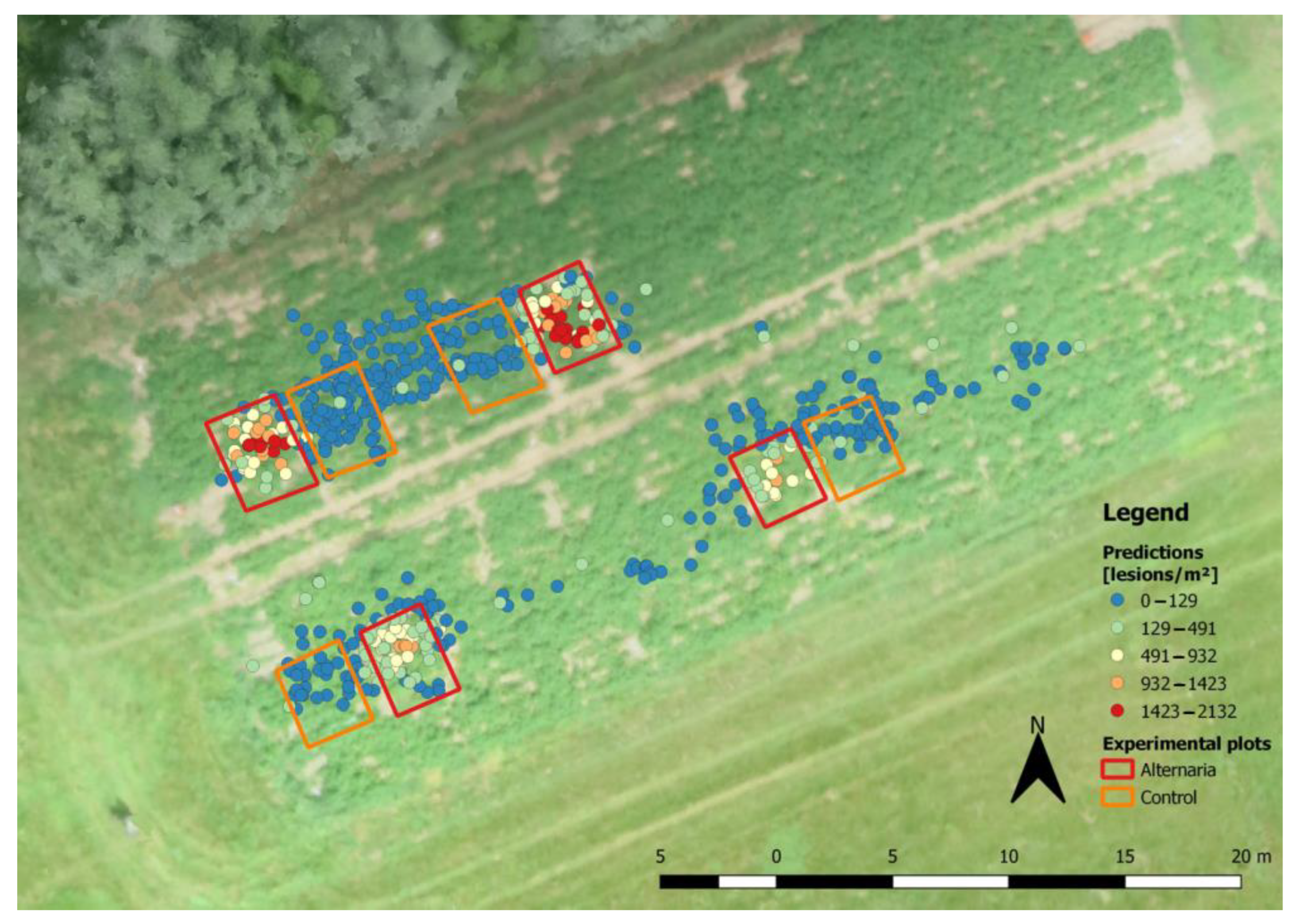
2.1. Field Trials
2.2. UAV Flights
2.3. Image Analysis Workflow
- : density estimate of lesion occurrence at pixel ij
- : the number of annotated lesions in the image
- : the distance to the annotated lesion
- : the bandwidth of the kernel density estimation
- : the number of pixels in an image
- : the ground truth value for the i-th pixel
- : the predicted value for the i-th pixel
3. Results
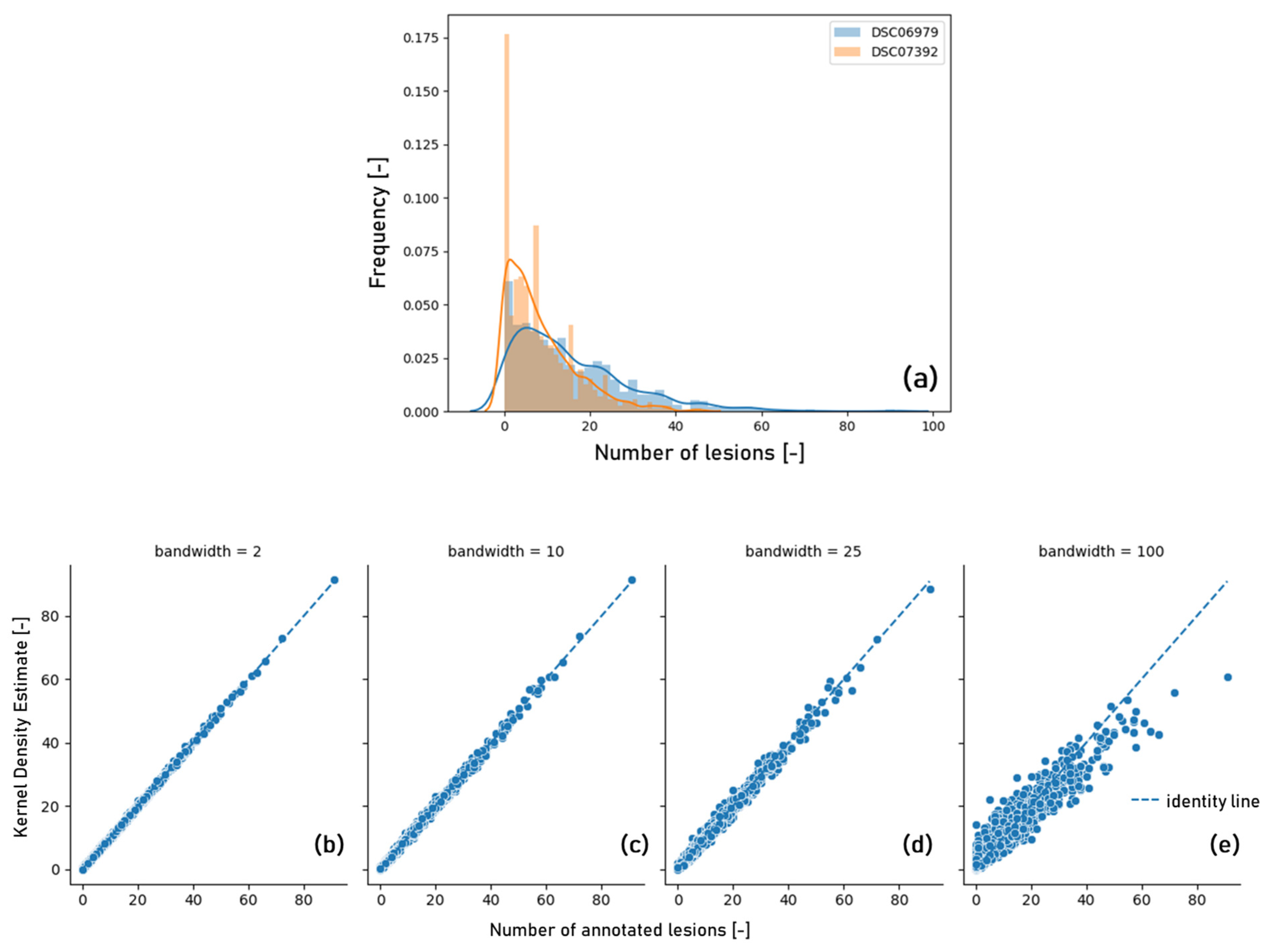
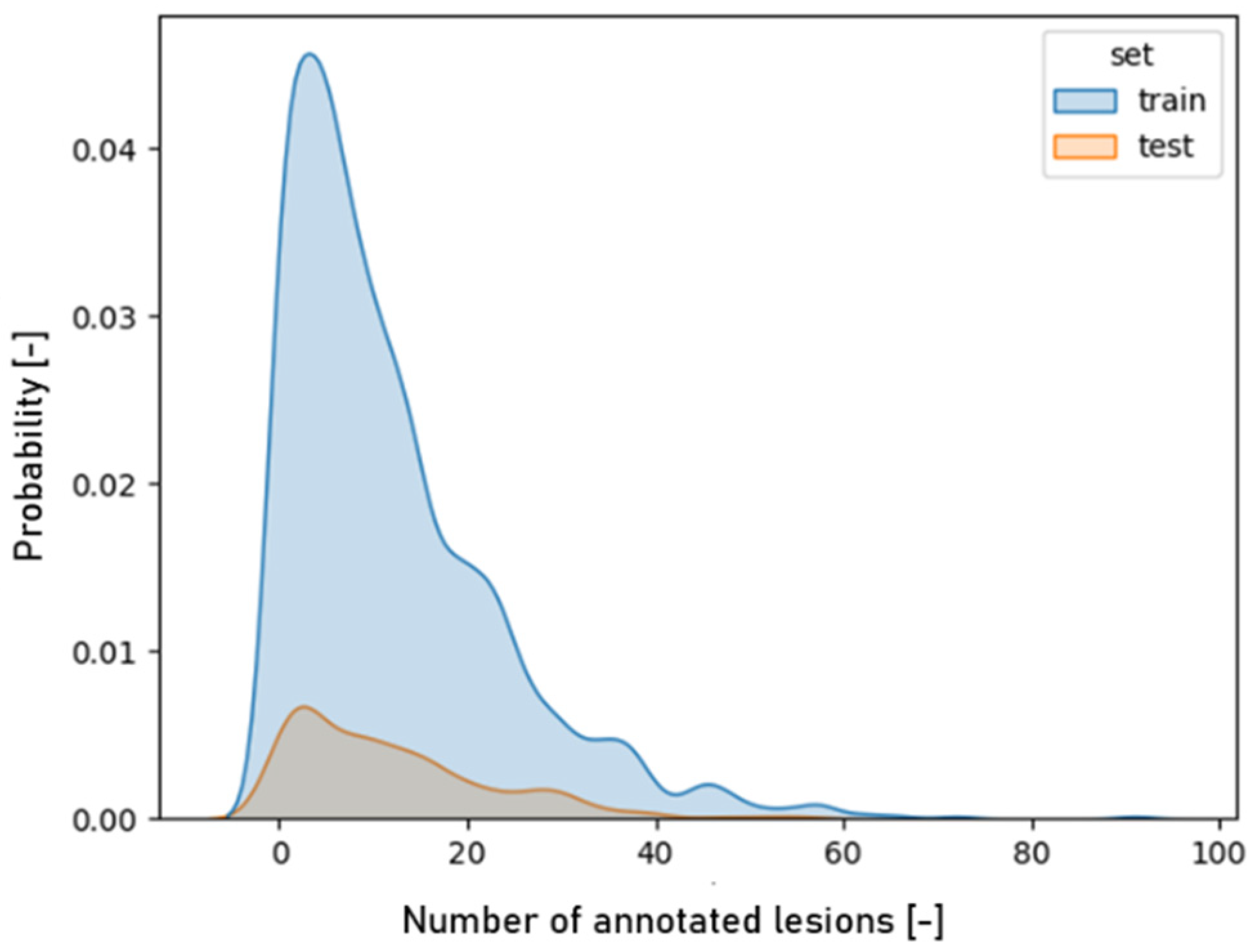
3.1. Image Characteristics
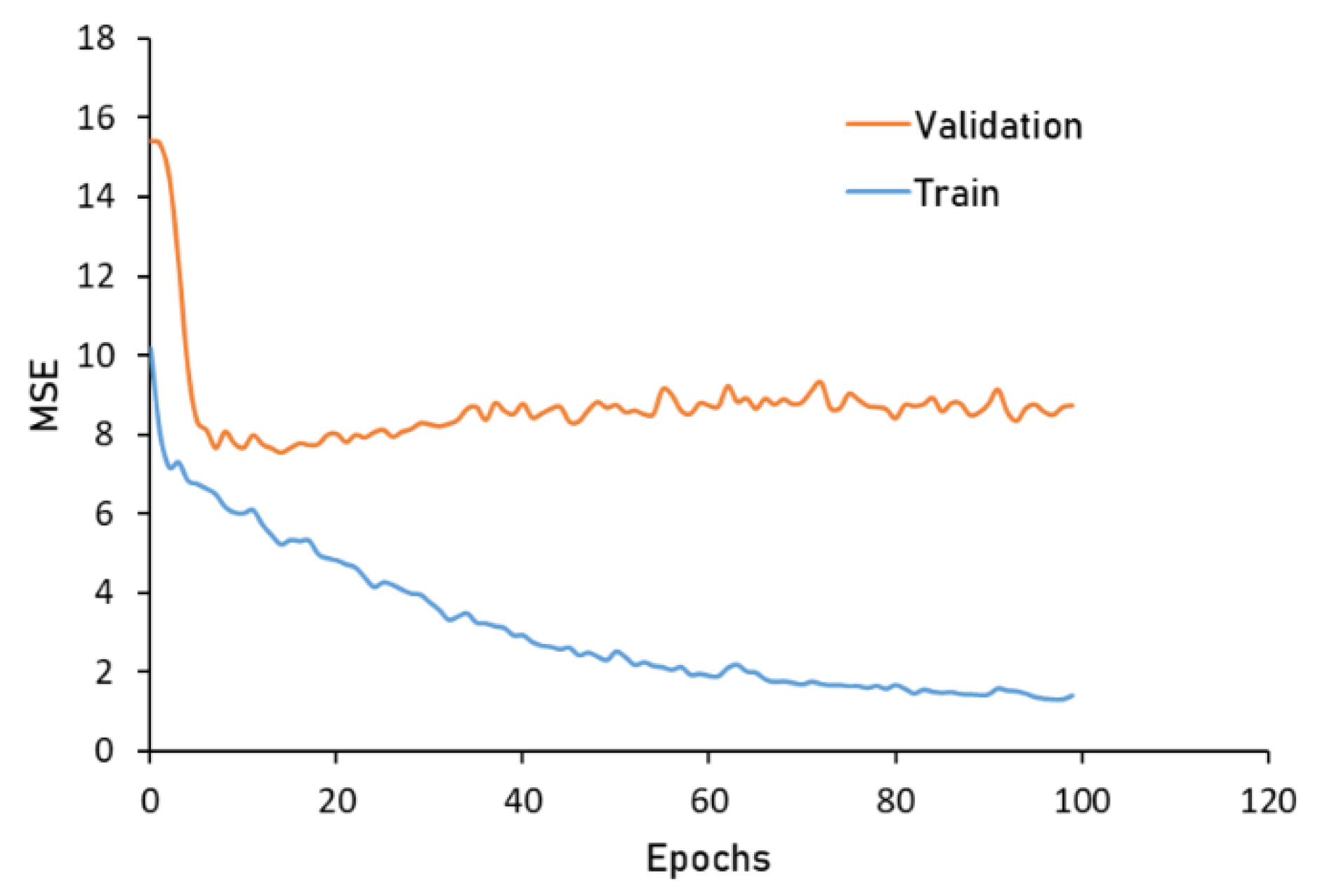
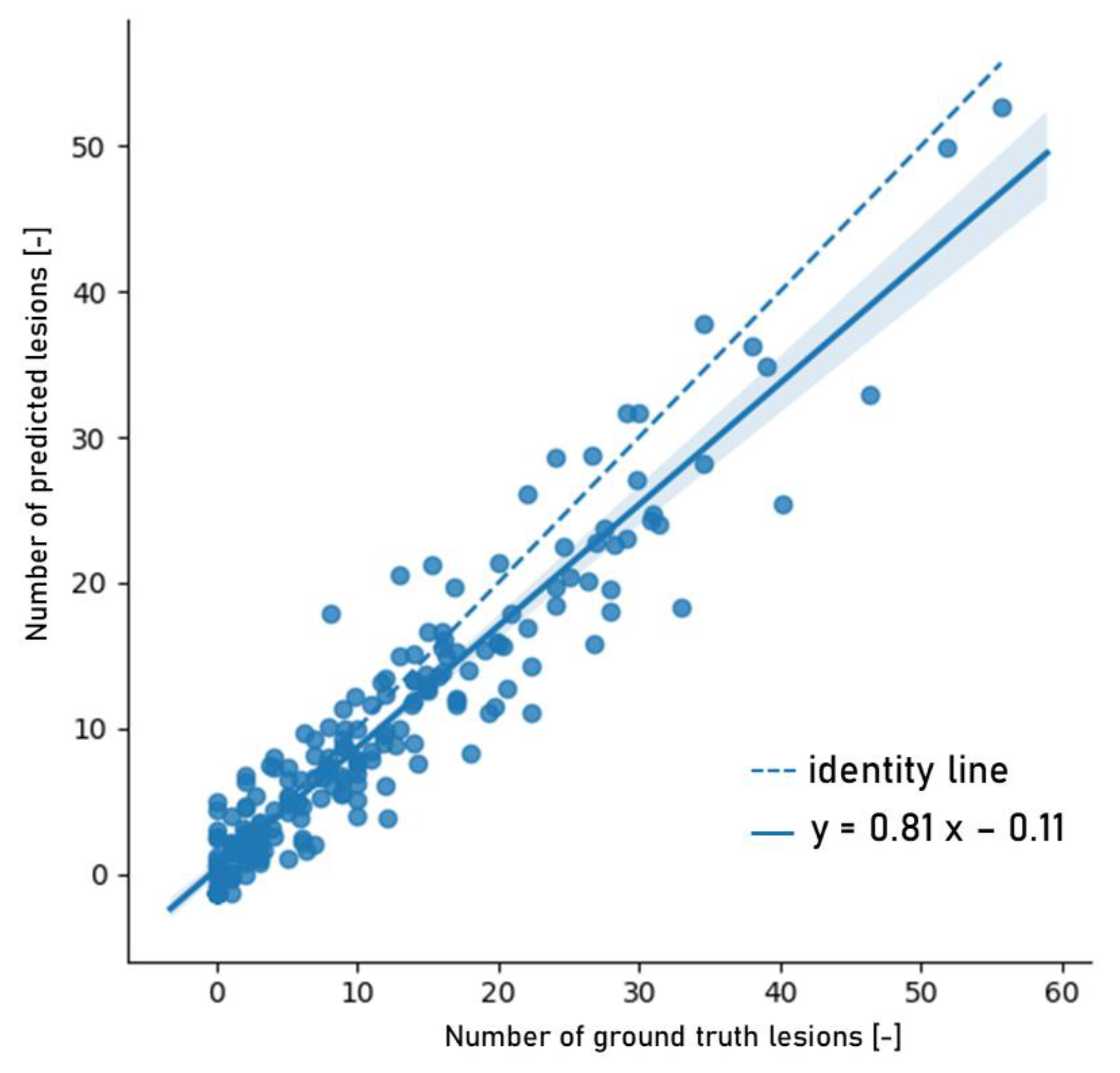
3.2. Model Analysis
4. Discussion
4.1. Image Characteristics
4.2. Model Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. Food and Agriculture Organization of the United Nations. 2022. Available online: https://www.fao.org/faostat/en/#home. (accessed on 20 November 2022).
- Horsfield, A.; Wicks, T.; Davies, K.; Wilson, D.; Paton, S. Effect of fungicide use strategies on the control of early blight (Alternaria solani) and potato yield. Australas. Plant Pathol. 2010, 39, 368–375. [Google Scholar] [CrossRef]
- Harrison, M.D.; Venette, J.R. Chemical control of potato early blight and its effect on potato yield. Am. Potato J. 1970, 47, 81–86. [Google Scholar] [CrossRef]
- Van der Waals, J.E.; Korsten, L.; Aveling, T.A.S. A review of early blight of potato. Afr. Plant Prot. 2001, 7, 91–102. [Google Scholar]
- Tsedaley, B. Review on early blight (Alternaria spp.) of potato disease and its management options. J. Biol. Agric. Healthc. 2014, 4, 191–199. [Google Scholar] [CrossRef] [Green Version]
- Leiminger, J.; Hausladen, H. Early blight: Influence of different varieties. PPO-Spec. Rep. 2007, 12, 195–203. [Google Scholar]
- American Phytopathological Society; Potato Association of America. Compendium of Potato Diseases; Hooker, W.J., Ed.; American Phytopathological Society: Saint Paul, MN, USA, 1981; ISBN 0890540276. [Google Scholar]
- Nasr-Esfahani, M. An IPM plan for early blight disease of potato Alternaria solani sorauer and A. alternata (Fries.) Keissler. Arch. Phytopathol. Plant Prot. 2020, 55, 785–796. [Google Scholar] [CrossRef]
- Zhang, N.; Yang, G.; Pan, Y.; Yang, X.; Chen, L.; Zhao, C. A review of advanced technologies and development for hyperspectral-based plant disease detection in the past three decades. Remote Sens. 2020, 12, 3188. [Google Scholar] [CrossRef]
- Mahlein, A. Present and Future Trends in Plant Disease Detection. Plant Dis. 2016, 100, 241–251. [Google Scholar] [CrossRef] [Green Version]
- Mahlein, A.; Kuska, M.T.; Behmann, J.; Polder, G.; Walter, A. Hyperspectral Sensors and Imaging Technologies in Phytopathology: State of the Art. Annu. Rev. Phytopathol. 2018, 56, 535–558. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, Y.; Pu, R.; Gonzalez-moreno, P.; Yuan, L. Monitoring plant diseases and pests through remote sensing technology: A review. Comput. Electron. Agric. 2019, 165, 104943. [Google Scholar] [CrossRef]
- Tetila, E.C.; Machado, B.B.; Menezes, G.K.; Oliveira, S.; Alvarez, M.; Amorim, W.P.; Alessandro, N.; Belete, D.S.; Gonçalves, G.; Pistori, H. Automatic Recognition of Soybean Leaf Diseases Using UAV Images and Deep Convolutional Neural Networks. IEEE Geosci. Remote Sens. Lett. 2019, 17, 903–907. [Google Scholar] [CrossRef]
- Zhang, X.; Han, L.; Dong, Y.; Shi, Y.; Huang, W.; Han, L.; González-Moreno, P.; Ma, H.; Ye, H.; Sobeih, T. A deep learning-based approach for automated yellow rust disease detection from high-resolution hyperspectral UAV images. Remote Sens. 2019, 11, 1554. [Google Scholar] [CrossRef]
- Maes, W.H.; Steppe, K. Perspectives for Remote Sensing with Unmanned Aerial Vehicles in Precision Agriculture. Trends Plant Sci. 2019, 24, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Dehkordi, R.H.; EL JARROUDI, M.; Kouadio, L.; Meersmans, J.; Beyer, M. Monitoring wheat leaf rust and stripe rust in winter wheat using high-resolution uav-based red-green-blue imagery. Remote Sens. 2020, 12, 3696. [Google Scholar] [CrossRef]
- Wiesner-Hanks, T.; Wu, H.; Stewart, E.; DeChant, C.; Kaczmar, N.; Lipson, H.; Gore, M.A.; Nelson, R.J. Millimeter-Level Plant Disease Detection From Aerial Photographs via Deep Learning and Crowdsourced Data. Front. Plant Sci. 2019, 10, 1550. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.P.; Yerudkar, A.; Mariani, V.; Iannelli, L.; Glielmo, L. A Bibliometric Review of the Use of Unmanned Aerial Vehicles in Precision Agriculture and Precision Viticulture for Sensing Applications. Remote Sens. 2022, 14, 1604. [Google Scholar] [CrossRef]
- Shafi, U.; Mumtaz, R.; García-nieto, J.; Hassan, S.A.; Zaidi, S.A.; Iqbal, N. Precision Agriculture Techniques and Practices: From consideration to application. Sensors 2019, 19, 3796. [Google Scholar] [CrossRef] [Green Version]
- Van De Vijver, R.; Mertens, K.; Heungens, K.; Somers, B.; Nuyttens, D.; Borra-serrano, I.; Lootens, P.; Roldán-ruiz, I.; Vangeyte, J.; Saeys, W. In-field detection of Alternaria solani in potato crops using hyperspectral imaging. Comput. Electron. Agric. 2020, 168, 105106. [Google Scholar] [CrossRef]
- Dutta, K.; Talukdar, D.; Bora, S.S. Segmentation of unhealthy leaves in cruciferous crops for early disease detection using vegetative indices and Otsu thresholding of aerial images. Measurement 2022, 189, 110478. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [Green Version]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Singh, D.; Jain, N.; Jain, P.; Kayal, P.; Kumawat, S.; Batra, N. PlantDoc: A dataset for visual plant disease detection. In Proceedings of the 7th ACM IKDD CoDS and 25th COMAD, Hyderabad, India, 5–7 January 2020; pp. 249–253. [Google Scholar] [CrossRef]
- Saleem, M.H.; Potgieter, J.; Arif, K.M. Plant disease detection and classification by deep learning. Plants 2019, 8, 32–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, A.M.; Bhaduri, J. A Deep Learning Enabled Multi-Class Plant Disease Detection Model Based on Computer Vision. Ai 2021, 2, 413–428. [Google Scholar] [CrossRef]
- Arsenovic, M.; Karanovic, M.; Sladojevic, S.; Anderla, A.; Stefanovic, D. Solving current limitations of deep learning based approaches for plant disease detection. Symmetry 2019, 11, 939. [Google Scholar] [CrossRef] [Green Version]
- Arnal Barbedo, J.G. Plant disease identification from individual lesions and spots using deep learning. Biosyst. Eng. 2019, 180, 96–107. [Google Scholar] [CrossRef]
- Kattenborn, T.; Eichel, J.; Fassnacht, F.E. Convolutional Neural Networks enable efficient, accurate and fine-grained segmentation of plant species and communities from high-resolution UAV imagery. Sci. Rep. 2019, 9, 17656. [Google Scholar] [CrossRef] [Green Version]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Proceedings of the Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1106–1114. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Singh, A.K.; Ganapathysubramanian, B.; Sarkar, S.; Singh, A. Deep Learning for Plant Stress Phenotyping: Trends and Future Perspectives Machine Learning in Plant Science. Trends Plant Sci. 2018, 23, 883–898. [Google Scholar] [CrossRef] [Green Version]
- Bouguettaya, A.; Zarzour, H.; Kechida, A.; Taberkit, A.M. Deep learning techniques to classify agricultural crops through UAV imagery: A review. Neural Comput. Appl. 2022, 34, 9511–9536. [Google Scholar] [CrossRef]
- QGIS Development Team. QGIS Geographic Information System 2019. Available online: https://qgis.org (accessed on 20 November 2022).
- Sam, D.B.; Surya, S.; Babu, R.V. Switching Convolutional Neural Network for Crowd Counting. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 5744–5752. [Google Scholar] [CrossRef] [Green Version]
- Pedregosa, F.; Michel, V.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Vanderplas, J.; Cournapeau, D.; Pedregosa, F.; Varoquaux, G.; et al. Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Zhang, Y.; Zhou, D.; Chen, S.; Gao, S.; Ma, Y. Single-image Crowd Counting via Multi-column Convolutional Neural Network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 589–597. [Google Scholar] [CrossRef]
- Ribera, J.; David, G.; Chen, Y.; Delp, E.J. Locating Objects Without Bounding Boxes. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 6479–6489. [Google Scholar]
- Zhou, X.; Wang, D.; Krähenbühl, P. Objects as Points. arXiv 2019, arXiv:1904.07850. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Yuan, Y.; Song, M.; Ding, Y.; Lin, F.; Liang, D. Use of Unmanned Aerial Vehicle Imagery and Deep Learning UNet to Extract Rice Lodging. Sensors 2019, 19, 3859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, K.; Chen, X.; Zhang, F.; Zhou, H.; Zhang, C. A field weed density evaluation method based on uav imaging and modified u-net. Remote Sens. 2021, 13, 310. [Google Scholar] [CrossRef]
- Wang, C.; Du, P.; Wu, H.; Li, J.; Zhao, C.; Zhu, H. A cucumber leaf disease severity classification method based on the fusion of DeepLabV3+ and U-Net. Comput. Electron. Agric. 2021, 189, 106373. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef] [Green Version]
- Yakubovskiy, P. Segmentation Models. In GitHub Repository; GitHub: San Francisco, CA, USA, 2019. [Google Scholar]
- Chollet, F. Keras; GitHub: San Francisco, CA, USA, 2015. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. arXiv 2015, arXiv:1603.04467. [Google Scholar]
- Tong, K.; Wu, Y.; Zhou, F. Recent advances in small object detection based on deep learning: A review. Image Vis. Comput. 2020, 97, 103910. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, D.; Yan, Y. Towards Efficient Detection for Small Objects via Attention-Guided Detection Network and Data Augmentation. Sensors 2022, 22, 7663. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. SSD: Single shot multibox detector. In Proceedings of the 14th European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; pp. 21–37. [Google Scholar] [CrossRef] [Green Version]
- Deng, Z.; Sun, H.; Zhou, S.; Zhao, J.; Lei, L.; Zou, H. Multi-scale object detection in remote sensing imagery with convolutional neural networks. ISPRS J. Photogramm. Remote Sens. 2018, 145, 3–22. [Google Scholar] [CrossRef]
- Li, J.; Wei, Y.; Liang, X.; Dong, J.; Xu, T. Attentive Contexts for Object Detection. IEEE Trans. Multimed. 2016, 19, 944–954. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Peng, C.; Yu, G.; Zhang, X.; Deng, Y.; Sun, J. DetNet: A Backbone network for Object. arXiv 2018, arXiv:1804.06215. [Google Scholar]
- Wackernagel, H. Ordinary Kriging. In Multivar. Geostatistics; Springer: Berlin/Heidelberg, Germany, 2003; pp. 79–88. [Google Scholar] [CrossRef]







| Red | Green | Blue | Density Map | |
|---|---|---|---|---|
| Mean | 0.72 | 0.27 | 0.079 | 0.00020 |
| Standard deviation | 0.22 | 0.12 | 0.063 | 0.00051 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van De Vijver, R.; Mertens, K.; Heungens, K.; Nuyttens, D.; Wieme, J.; Maes, W.H.; Van Beek, J.; Somers, B.; Saeys, W. Ultra-High-Resolution UAV-Based Detection of Alternaria solani Infections in Potato Fields. Remote Sens. 2022, 14, 6232. https://doi.org/10.3390/rs14246232
Van De Vijver R, Mertens K, Heungens K, Nuyttens D, Wieme J, Maes WH, Van Beek J, Somers B, Saeys W. Ultra-High-Resolution UAV-Based Detection of Alternaria solani Infections in Potato Fields. Remote Sensing. 2022; 14(24):6232. https://doi.org/10.3390/rs14246232
Chicago/Turabian StyleVan De Vijver, Ruben, Koen Mertens, Kurt Heungens, David Nuyttens, Jana Wieme, Wouter H. Maes, Jonathan Van Beek, Ben Somers, and Wouter Saeys. 2022. "Ultra-High-Resolution UAV-Based Detection of Alternaria solani Infections in Potato Fields" Remote Sensing 14, no. 24: 6232. https://doi.org/10.3390/rs14246232
APA StyleVan De Vijver, R., Mertens, K., Heungens, K., Nuyttens, D., Wieme, J., Maes, W. H., Van Beek, J., Somers, B., & Saeys, W. (2022). Ultra-High-Resolution UAV-Based Detection of Alternaria solani Infections in Potato Fields. Remote Sensing, 14(24), 6232. https://doi.org/10.3390/rs14246232







