Improved Object-Based Mapping of Aboveground Biomass Using Geographic Stratification with GEDI Data and Multi-Sensor Imagery
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Study Area
2.2. Data
2.2.1. Ground-Sampled Measurements
2.2.2. Pre-Processing of Multi-Sensor Data
2.3. Methods
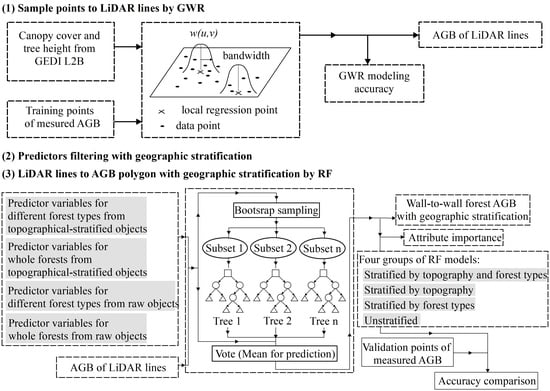
2.3.1. GWR Modeling for GEDI-Derived AGB Lines
2.3.2. Filtering Predictors for Different Forest Types from Topographically Stratified Objects
2.3.3. Prediction Stratified or Raw AGB Polygons by RF
3. Results
3.1. GEDI LiDAR-Extracted AGB Lines Using GWR
3.2. Predictor Variables for Different Forest Types from Topographically Stratified Objects
3.3. RF Models for AGB Prediction
3.4. Forest AGB Map Predicted by Geographic Stratification
4. Discussion
4.1. Role of Topographical Stratification in AGB Estimation
4.2. Influence of Stratification by Forest Type for AGB Estimation
4.3. Uncertainty of Geographic Stratification-Based AGB Mapping
5. Conclusions
- (1)
- The geographic stratification approach precisely estimated the biomass of heterogeneous forests, and improved the accuracy by 34.79% more than the unstratified process. The stratification of forest types further increased the mapped AGB accuracy compared to that of topography. Topographical data with a finer spatiotemporal resolution and forests with distinguished vertical zonality may enhance this contribution.
- (2)
- The relationships between multi-sensor variables and AGB varied within the different approaches of geographic stratification. Generally, vegetation indices from S2 MSI, texture features from VV backscatters, and elevation were the most important predictors in AGB modeling. Topographical stratification greatly influenced the predictors’ contributions to AGB mapping in mixed broadleaf–conifer and broad-leaved forests, but only slightly impacted coniferous forests. Optical variables were predominant for deciduous forests, while for evergreen forests, the SAR indices outweighed the other predictors.
- (3)
- The mapped values from 8.41 and 504.82 Mg/ha were approximate to the ground-observed forest AGB. The western and northern deciduous broad-leaved forests, with elevations of 600–800 m, had the largest AGB values. The smallest values were distributed in deciduous coniferous forests with an altitude above 800 m.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luo, K.; Wei, Y.; Du, J.; Liu, L.; Luo, X.; Shi, Y.; Pei, X.; Lei, N.; Song, C.; Li, J.; et al. Machine learning-based estimates of aboveground biomass of subalpine forests using Landsat 8 OLI and Sentinel-2B images in the Jiuzhaigou National Nature Reserve, Eastern Tibet Plateau. J. Forestry Res. 2022, 33, 1329–1340. [Google Scholar] [CrossRef]
- Harris, N.L.; Gibbs, D.A.; Baccini, A.; Birdsey, R.A.; de Bruin, S.; Farina, M.; Fatoyinbo, L.; Hansen, M.C.; Herold, M.; Houghton, R.A.; et al. Global maps of twenty-first century forest carbon fluxes. Nat. Clim. Chang. 2021, 11, 234–240. [Google Scholar] [CrossRef]
- Chopping, M.; Wang, Z.; Schaaf, C.; Bull, M.A.; Duchesne, R.R. Forest aboveground biomass in the southwestern United States from a MISR multi-angle index, 2000–2015. Remote Sens. Environ. 2022, 275, 112964. [Google Scholar] [CrossRef]
- Liang, M.; Duncanson, L.; Silva, J.A.; Sedano, F. Quantifying aboveground biomass dynamics from charcoal degradation in Mozambique using GEDI Lidar and Landsat. Remote Sens. Environ. 2023, 284, 113367. [Google Scholar] [CrossRef]
- Qin, Y.; Xiao, X.; Wigneron, J.P.; Ciais, P.; Canadell, J.G.; Brandt, M.; Li, X.; Fan, L.; Wu, X.; Tang, H.; et al. Large loss and rapid recovery of vegetation cover and aboveground biomass over forest areas in Australia during 2019–2020. Remote Sens. Environ. 2022, 224, 1–11. [Google Scholar] [CrossRef]
- Cartus, O.; Santoro, M.; Kellndorfer, J. Mapping forest aboveground biomass in the Northeastern United States with ALOS PALSAR dual-polarization L-band. Remote Sens. Environ. 2012, 124, 466–478. [Google Scholar] [CrossRef]
- Santoro, M.; Cartus, O.; Fransson, J.E.S. Integration of allometric equations in the water cloud model towards an improved retrieval of forest stem volume with L-band SAR data in Sweden. Remote Sens. Environ. 2021, 253, 112235. [Google Scholar] [CrossRef]
- Cutler, M.E.J.; Boyd, D.S.; Foody, G.M.; Vetrivel, A. Estimating tropical forest biomass with a combination of SAR image texture and Landsat TM data: An assessment of predictions between regions. ISPRS J. Photogramm. 2012, 70, 66–77. [Google Scholar] [CrossRef]
- Poorazimy, M.; Shataee, S.; McRoberts, R.E.; Mohammadi, J. Integrating airborne laser scanning data, space-borne radar data and digital aerial imagery to estimate aboveground carbon stock in Hyrcanian forests, Iran. Remote Sens. Environ. 2020, 240, 111669. [Google Scholar] [CrossRef]
- Clark, M.L.; Roberts, D.A.; Ewel, J.J.; Clark, D.B. Estimation of tropical rain forest aboveground biomass with small-footprint lidar and hyperspectral sensors. Remote Sens. Environ. 2011, 115, 2931–2942. [Google Scholar] [CrossRef]
- Mahoney, M.J.; Johnson, L.K.; Bevilacqua, E.; Beier, C.M. Filtering ground noise from LiDAR returns produces inferior models of forest aboveground biomass in heterogenous landscapes. GISci. Remote Sens. 2022, 59, 1266–1280. [Google Scholar] [CrossRef]
- Su, Y.; Guo, Q.; Xue, B.; Hu, T.; Alvarez, O.; Tao, S.; Fang, J. Spatial distribution of forest aboveground biomass in China: Estimation through combination of spaceborne lidar, optical imagery, and forest inventory data. Remote Sens. Environ. 2016, 173, 187–199. [Google Scholar] [CrossRef]
- Nandy, S.; Srinet, R.; Padalia, H. Mapping forest height and aboveground biomass by integrating ICESat-2, Sentinel-1 and Sentinel-2 data using random forest algorithm in northwest Himalayan foothills of India. Geophys. Res. Lett. 2021, 48, e2021GL093799. [Google Scholar] [CrossRef]
- Shendryk, Y. Fusing GEDI with earth observation data for large area aboveground biomass mapping. Int. J. Appl. Earth Obs. 2022, 115, 103108. [Google Scholar] [CrossRef]
- Silveira, E.M.O.; Radeloff, V.C.; Martinuzzi, S.; Pastur, G.J.M.; Bono, J.; Politi, N.; Lizarraga, L.; Rivera, L.O.; Ciuffoli, L.; Rosas, Y.M.; et al. Nationwide native forest structure maps for Argentina based on forest inventory data, SAR Sentinel-1 and vegetation metrics from Sentinel-2 imagery. Remote Sens. Environ. 2023, 285, 113391. [Google Scholar] [CrossRef]
- Tamiminia, H.; Salehi, B.; Mahdianpari, M.; Beier, C.M.; Johnson, L. Mapping two decades of New York State forest aboveground biomass change using remote sensing. Remote Sens. 2022, 14, 4097. [Google Scholar] [CrossRef]
- Silveira, E.M.O.; Silva, S.H.G.; Acerbi-Junior, F.W.; Carvalho, M.C.; Carvalho, L.M.T.; Scolforo, J.R.S.; Wulder, M.A. Object-based random forest modeling of aboveground forest biomass outperforms a pixel-based approach in a heterogeneous and mountain tropical environment. Int. J. Appl. Earth Obs. 2019, 78, 175–188. [Google Scholar]
- Fassnacht, F.E.; Hartig, F.; Latifi, H.; Berger, C.; Hernández, J.; Corvalán, P.; Koch, B. Importance of sample size, data type and prediction method for remote sensing-based estimations of aboveground forest biomass. Remote Sens. Environ. 2014, 154, 102–114. [Google Scholar] [CrossRef]
- Narine, L.L.; Popescu, S.; Neuenschwander, A.; Zhou, T.; Srinivasan, S.; Harbeck, K. Estimating aboveground biomass and forest canopy cover with simulated ICESat-2 data. Remote Sens. Environ. 2019, 224, 1–11. [Google Scholar] [CrossRef]
- Zhou, G.; Yi, G.; Tang, X.; Wen, Z.; Liu, C.; Kuang, Y.; Wang, W. Carbon Stock of Forest Ecosystems in China—Biomass Equations; Science Press: Beijing, China, 2018. [Google Scholar]
- Huang, W.; Dolan, K.; Swatantran, A.; Johnson, K.; Tang, H.; O’Neil-Dunne, J.; Dubayah, R.; Hurtt, G. High-resolution mapping of aboveground biomass for forest carbon monitoring system in the Tri-State region of Maryland, Pennsylvania and Delaware, USA. Environ. Res. Lett. 2019, 14, 095002. [Google Scholar] [CrossRef]
- Jiang, F.; Deng, M.; Tang, J.; Fu, L.; Sun, H. Integrating spaceborne LiDAR and Sentinel-2 images to estimate forest aboveground biomass in Northern China. Carbon Bal. Manag. 2022, 17, 12. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.A.; Hamamura, C.; Valbuena, R.; Hancock, S.; Cardil, A.; Broadbent, E.N.; Almeida, D.R.A.; Silva Junior, C.H.L.; Klauberg, C. rGEDI: NASA’s Global Ecosystem Dynamics Investigation (GEDI) Data Visualization and Processing. 2020. version 0.1.2. 2020. Available online: https://CRAN.R-project.org/package=rGEDI (accessed on 1 April 2020).
- Hird, J.N.; DeLancey, E.R.; McDermid, G.J.; Kariyeva, J. Google Earth Engine, open-access satellite data, and machine learning in support of large-area probabilistic wetland mapping. Remote Sens. 2017, 9, 1315. [Google Scholar] [CrossRef]
- Fernandez-Carrillo, A.; McCaw, L.; Tanase, M.A. Estimating prescribed fire impacts and post-fire tree survival in eucalyptus forests of Western Australia with L-band SAR data. Remote Sens. Environ. 2019, 224, 133–144. [Google Scholar] [CrossRef]
- Liu, Y.; Gong, W.; Xing, Y.; Hu, X.; Gong, J. Estimation of the forest stand mean height and aboveground biomass in Northeast China using SAR Sentinel-1B, multispectral Sentinel-2A, and DEM imagery. ISPRS J. Photogramm. 2019, 151, 277–289. [Google Scholar] [CrossRef]
- Fremout, T.; Cobián-De Vinatea, J.; Thomas, E.; Huaman-Zambrano, W.; Salazar-Villegas, M.; la Fuente, D.L.; Bernardino, P.N.; Atkinson, R.; Csaplovics, E.; Muys, B. Site-specific scaling of remote sensing-based estimates of woody cover and aboveground biomass for mapping long-term tropical dry forest degradation status. Remote Sens. Environ. 2022, 276, 113040. [Google Scholar] [CrossRef]
- Castillo, J.A.A.; Apan, A.A.; Maraseni, T.N.; Salmo, S.G., III. Estimation and mapping of above-ground biomass of mangrove forests and their replacement land uses in the Philippines using Sentinel imagery. ISPRS J. Photogramm. 2017, 134, 70–85. [Google Scholar] [CrossRef]
- Forkuor, G.; Zoungrana, J.B.B.; Dimobe, K.; Ouattara, B.; Vadrevu, K.P.; Tondoh, J.E. Above-ground biomass mapping in West African dryland forest using Sentinel-1 and 2 datasets—A case study. Remote Sens. Environ. 2020, 236, 111496. [Google Scholar] [CrossRef]
- Kearney, S.P.; Porensky, L.M.; Augustine, D.J.; Gaffney, R.; Derner, J.D. Monitoring standing herbaceous biomass and thresholds in semiarid rangelands from harmonized Landsat 8 and Sentinel-2 imagery to support within-season adaptive management. Remote Sens. Environ. 2022, 271, 112907. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Guo, Z. Estimation of tree height and aboveground biomass of coniferous forests in North China using stereo ZY-3, multispectral Sentinel-2, and DEM data. Ecol. Indic. 2021, 126, 107645. [Google Scholar] [CrossRef]
- Bell, D.M.; Gregory, M.J.; Kane, V.; Kane, J.; Kennedy, R.E.; Roberts, H.M.; Yang, Z. Multiscale divergence between Landsat- and lidar-based biomass mapping is related to regional variation in canopy cover and composition. Carbon Bal. Manag. 2018, 13, 15. [Google Scholar] [CrossRef]
- Yang, Q.; Su, Y.; Hu, T.; Jin, S.; Liu, X.; Niu, C.; Liu, Z.; Kelly, M.; Wei, J.; Guo, Q. Allometry-based estimation of forest aboveground biomass combining LiDAR canopy height attributes and optical spectral indexes. For. Ecosyst. 2022, 9, 100059. [Google Scholar] [CrossRef]
- Brunsdon, C.; Fotheringham, A.S.; Charlton, M.E. Geographically weighted regression: A method for exploring spatial nonstationarity. Geogr. Anal. 1996, 28, 281–298. [Google Scholar] [CrossRef]
- Fotheringham, A.S.; Brunsdon, C.; Charlton, M.E. Geographically Weighted Regression: The Analysis of Spatially Varying Relationships; Wiley: Chichester, UK, 2002. [Google Scholar]
- Shin, J.; Temesgen, H.; Strunk, J.L.; Hilker, T. Comparing modeling methods for predicting forest attributes using LiDAR metrics and ground measurements. Can. J. Remote Sens. 2016, 42, 739–765. [Google Scholar] [CrossRef]
- Nakaya, T.; Charlton, M.; Lewis, P.; Brunsdon, C.; Yao, J.; Fotheringham, S. GWR4 User Manual, Windows Application for Geographically Weighted Regression Modelling; Ritsumeikan University: Kyoto, Japan, 2014. [Google Scholar]
- Zhang, C.; Denka, S.; Cooper, H.; Mishra, D.R. Quantification of sawgrass marsh aboveground biomass in the coastal Everglades using object-based ensemble analysis and Landsat data. Remote Sens. Environ. 2018, 204, 366–379. [Google Scholar] [CrossRef]
- Millard, K.; Richardson, M.C. On the importance of training data sample selection in RF classification: A case study in peatland ecosystem mapping. Remote Sens. 2015, 7, 8489–8515. [Google Scholar] [CrossRef]
- Xu, C.; Manley, B.; Morgenroth, J. Evaluation of modeling approaches in predicting forest volume and stand age for small-scale plantation forests in New Zealand with RapidEye and LiDAR. Int. J. Appl. Earth Obs. 2018, 73, 386–396. [Google Scholar]
- Lu, D.; Chen, Q.; Wang, G.; Liu, L.; Li, G.; Moran, E. A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2016, 9, 63–105. [Google Scholar] [CrossRef]
- Zhao, Q.; Yu, S.; Zhao, F.; Tian, L.; Zhao, Z. Comparison of machine learning algorithms for forest parameter estimations and application for forest quality assessments. Forest Ecol. Manag. 2019, 434, 224–234. [Google Scholar] [CrossRef]
- Wittke, S.; Yu, X.W.; Karjalainen, M.; Hyyppä, J.; Puttonen, E. Comparison of two-dimensional multitemporal Sentinel-2 data with three dimensional remote sensing data sources for forest inventory parameter estimation over a boreal forest. Int. J. Appl. Earth Obs. 2019, 76, 167–178. [Google Scholar] [CrossRef]
- Cao, J.; Liu, K.; Zhuo, L.; Liu, L.; Zhu, Y.; Peng, L. Combining UAV-based hyperspectral and LiDAR data for mangrove species classification using the rotation forest algorithm. Int. J. Appl. Earth Obs. 2021, 102, 102414. [Google Scholar] [CrossRef]
- Naidoo, L.; Mathieu, R.; Main, R.; Kleynhans, W.; Wessels, K.; Asner, G.; Leblon, B. Savannah woody structure modeling and mapping using multi-frequency (X-, C- and L-band) Synthetic Aperture Radar data. ISPRS J. Photogramm. 2015, 105, 234–250. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, J.; Liang, S.; Li, X.; Liu, J. A stacking ensemble algorithm for improving the biases of forest aboveground biomass estimations from multiple remotely sensed datasets. GISci. Remote Sens. 2022, 59, 234–249. [Google Scholar] [CrossRef]
- Nevalainen, O.; Honkavaara, E.; Tuominen, S.; Viljanen, N.; Hakala, T.; Yu, X.; Hyyppä, J.; Saari, H.; Pölönen, I.; Imai, N.N.; et al. Individual tree detection and classification with UAV-based photogrammetric point clouds and hyperspectral imaging. Remote Sens. 2017, 9, 185. [Google Scholar] [CrossRef]
- Schwaab, J.; Davin, E.L.; Bebi, P.; Duguay-Tetzlaff, A.; Waser, L.T.; Haeni, M.; Meier, R. Increasing the broad-leaved tree fraction in European forests mitigates hot temperature extremes. Sci. Rep. 2020, 10, 14153. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Veiga, P.; Quegan, S.; Carreiras, J.; Persson, H.J.; Fransson, J.E.S.; Hoscilo, A.; Ziółkowski, D.; Stereńczak, K.; Lohberger, S.; Stängel, M.; et al. Forest biomass retrieval approaches from earth observation in different biomes. Int. J. Appl. Earth Obs. 2019, 77, 53–68. [Google Scholar] [CrossRef]
- Chen, L.; Ren, C.; Zhang, B.; Wang, Z.; Liu, M.; Man, W.; Liu, J. Improved estimation of forest stand volume by the integration of GEDI LiDAR data and multi-sensor imagery in the Changbai Mountains Mixed Forests Ecoregion (CMMFE), Northeast China. Int. J. Appl. Earth Obs. 2021, 100, 102326. [Google Scholar] [CrossRef]
- Le Maire, G.; François, C.; Soudani, K.; Berveiller, D.; Pontailler, J.-Y.; Bréda, N.; Genet, H.; Davi, H.; Dufrêne, E. Calibration and validation of hyperspectral indices for the estimation of broadleaved forest leaf chlorophyll content, leaf mass per area, leaf area index and leaf canopy biomass. Remote Sens. Environ. 2008, 112, 3846–3864. [Google Scholar] [CrossRef]
- Mao, P.; Qin, L.; Hao, M.; Zhao, W.; Luo, J.; Qiu, X.; Xu, L.; Xiong, Y.; Ran, Y.; Yan, C.; et al. An improved approach to estimate above-ground volume and biomass of desert shrub communities based on UAV RGB images. Ecol. Indic. 2021, 125, 107494. [Google Scholar] [CrossRef]
- Galeana-Pizaña, J.M.; López-Caloca, A.; López-Quiroz, P.; Silván-Cárdenas, J.L.; Couturier, S. Modeling the spatial distribution of above-ground carbon in Mexican coniferous forests using remote sensing and a geostatistical approach. Int. J. Appl. Earth Obs. 2014, 30, 179–189. [Google Scholar] [CrossRef]
- MOF (Ministry of Forestry). Standards for Forestry Resource Survey. China; Forestry Publisher: Beijing, China, 1982. [Google Scholar]
- Chen, L.; Ren, C.; Bao, G.; Zhang, B.; Wang, Z.; Liu, M.; Man, W.; Liu, J. Improved object-based estimation of forest aboveground biomass by integrating LiDAR data from GEDI and ICESat-2 with multi-sensor images in a heterogeneous mountainous region. Remote Sens. 2022, 14, 2743. [Google Scholar] [CrossRef]








| Tree Species/Family/Types | Trunk | Branch | Leaf |
|---|---|---|---|
| Betula platyphylla Suk. | 0.0789 × (D2 × H)0.8607 | 0.0090 × (D2 × H)0.8742 | 0.0051 × (D2 × H)0.7552 |
| Betula dahurica Pall. | 0.0842 × (D2 × H)0.7965 | 0.0033 × (D2 × H)1.0630 | 0.0035 × (D2 × H)0.8603 |
| Pinus koraiensis Siebold et Zuccarini | 0.0204 × (D2 × H)0.9822 | 0.0119 × (D2 × H)0.7457 | 0.0594 × (D2 × H)0.5125 |
| Quercus mongolica Fisch. ex Ledeb. | 0.1131 × (D2 × H)0.8631 | 0.0049 × (D2 × H)1.1016 | 0.0156 × (D2 × H)0.7295 |
| Populus davidiana Dode | 0.1281 × (D2 × H)0.6952 | 0.0234 × (D2 × H)0.7496 | 0.0103 × (D2 × H)0.8309 |
| Pinus sylvestris var. mongolica Litv. | 0.0995 × (D2 × H)0.7656 | 0.0464 × (D2 × H)0.6778 | 0.0422 × (D2 × H)0.6372 |
| Picea asperata Mast./Abies fabri (Mast.) Craib/Abies nephrolepis (Trautv.) Maxim. | 0.0408 × (D2 × H)0.9020 | 0.0953 × (D2 × H)0.6714 | 0.1049 × (D2 × H)0.6249 |
| Betula | 0.1040 × (D2 × H)0.7926 | 0.0087 × (D2 × H)0.8855 | 0.0064 × (D2 × H)0.7453 |
| Larix gmelinii (Rupr.) Kuzen. | 0.0242 × (D2 × H)0.9445 | 0.0040 × (D2 × H)0.9272 | 0.0091 × (D2 × H)0.7482 |
| Populus L. | 0.0340 × (D2 × H)0.9160 | 0.0090 × (D2 × H)0.9150 | 0.0060 × (D2 × H)0.7890 |
| Other broad-leaved trees | 0.0256 × (D2 × H)0.9553 | 0.0119 × (D2 × H)0.7566 | 0.0477 × (D2 × H)0.5390 |
| Other coniferous trees | 0.1295 × (D2 × H)0.8076 | 0.0062 × (D2 × H)0.9587 | 0.0139 × (D2 × H)0.7245 |
| Mixed broadleaf–conifer forests | 0.0768 × (D2 × H)0.8563 | 0.0085 × (D2 × H)0.8707 | 0.0219 × (D2 × H)0.6526 |
| Source | Level | Spatial Resolution | Date | Compositions |
|---|---|---|---|---|
| GEDI | 2B | 25 m | 20190507, 0514, 0521, 0526, 0620, 0627, 0716, 0821, 0829, 0912, 0921 | T00592, 04708, 04555, 01522, 01675, 01862, 04368, 00286, 04062, 00745, 00099 |
| A2 | Yearly mosaic | 25 m | 2019 | N44E128 |
| S1 | Ground Range Detected (GRD) scenes | 10 m | 20190504, 0511, 0523, 0604, 0616, 0628, 0710, 0722, 0803, 0815, 0827, 0901, 0908, 0913, 0920 | S1A_030CEC_BAD6, 3104D_D943, 315C5_02E2, 31B36_1AB8, 3207F_6ECF, 325B7_5070, 32B0B_A562, 33052_8A6E, 335A7_6B96, 33B72_1513, 34189_C444, 34415_C219, 3479D_8BE2/597F, 34A27_6CE3, 34DA7_F7A5 |
| 20190503, 0508, 0515, 0520, 0527, 0601, 0608, 0613, 0620, 0625, 0702, 0714, 0719, 0726, 0731, 0807, 0812, 0819, 0831, 0905, 0912, 0917, 0924, 0929 | S1B_1E41C_0156, 1E671_A592, 1E9A3_82B7, 1EBD2_6809, 1EEFF_609F, 1F128_409B, 1F438_BCD5, 1F65C_83E3, 1F96F_D10F, 1FB87_7017, 1FE9A_FD21, 203C2_FF18, 205CF_82EE, 208D7_1E72, 20B01_ED20, 20E22_CBD8, 2105F_16BB, 21398_3093, 21909_FCDD, 21B40_B939, 21E81_83FB, 220BA_9277, 223E8_5B7B, 2261D_6C27 | |||
| S2 | 2A, orthorectified atmospherically corrected surface reflectance | 10 m | 20190503, 0506, 0513, 0516, 0523, 0526, 0602, 0605, 0612, 0615, 0622, 0625, 0702, 0705, 0712, 0715, 0722, 0725, 0801, 0804, 0811, 0814, 0821, 0824, 0831, 0903, 0910, 0913, 0920, 0923, 0930 | There is one image on each date as S2A_T52TDP. |
| 20190501, 0508, 0511, 0518, 0528, 0531, 0607, 0610, 0617, 0620, 0627, 0630, 0707, 0710, 0717, 0720, 0727, 0730, 0806, 0809, 0816, 0819, 0826, 0829, 0905, 0908, 0915, 0918, 0925, 0928 | There is one image on each date as S2B_T52TDP. | |||
| A1 | DSM | 30 m | Derived from A1 SAR data from 2006 to 2011 | N043E128 |
| Images | Variables | Explanation | |
|---|---|---|---|
| A2 mosaic | Backscatter | HH | Normalized backscatter coefficient of the horizontal transmit-horizontal channel in dB |
| HV | Normalized backscatter coefficient of the vertical transmit-vertical channel in dB | ||
| RFDI | Radar forest degradation index | ||
| V/H_L | HV/HH | ||
| S1 mosaic | Backscatter | VV | Normalized backscatter coefficient of the vertical transmit-vertical channel in dB |
| VH | Normalized backscatter coefficient of the vertical transmit-horizontal channel in dB | ||
| NP | Normalized polarization | ||
| V/H_C | VV/VH | ||
| Texture | VV/VH_CON | Contrast | |
| VV/VH_DIS | Dissimilarity | ||
| VV/VH_HOM | Homogeneity | ||
| VV/VH_ASM | Angular second moment | ||
| VV/VH_ENE | Energy | ||
| VV/VH_MAX | Maximum probability | ||
| VV/VH_ENT | Entropy | ||
| VV/VH_MEA | Gray-level co-occurrence matrix (GLCM) mean | ||
| VV/VH_VAR | GLCM variance | ||
| VV/VH_COR | GLCM correlation | ||
| S2 mosaic | Multispectral bands | B2 | Blue |
| B3 | Green | ||
| B4 | Red | ||
| B5 | Red edge | ||
| B6 | Red edge | ||
| B7 | Red edge | ||
| B8 | Near-infrared | ||
| B8a | Near-infrared | ||
| B11 | Short-wave infrared | ||
| B12 | Short-wave infrared | ||
| Vegetation indices | RVI | Ratio vegetation index | |
| DVI | Difference vegetation index | ||
| NDVI | Normalized difference vegetation index | ||
| EVI | Enhanced vegetation index | ||
| S2REP | Sentinel-2 red-edge position index | ||
| REIP | Red-edge infection point index | ||
| SAVI | Soil adjusted vegetation index | ||
| MTCI | Meris terrestrial chlorophyll index | ||
| MCARI | Modified chlorophyll absorption ratio index | ||
| DSM | Topographic indicators | H | Elevation |
| β | Slope | ||
| A | Aspect | ||
| M | Surface roughness | ||
| TWI | Topographic wetness index | ||
| SPI | Stream power index | ||
| Variables | Minimum | Maximum | Mean | Medium | SD |
|---|---|---|---|---|---|
| C | 0.001 | 0.99 | 0.60 | 0.73 | 0.31 |
| Ht (m) | 1.35 | 54.39 | 18.17 | 19.66 | 9.62 |
| AGB (Mg/ha) | 4.51 | 544.90 | 137.33 | 142.53 | 38.22 |
| Group | ME | RMSE | R2 | RIRMSE (%) | ||
|---|---|---|---|---|---|---|
| Mg/ha | % | Mg/ha | % | |||
| Stratified by topography and forest types | 12.29 | 8.74 | 32.72 | 23.28 | 0.92 | 34.79 |
| Stratified by topography | 14.15 | 10.07 | 48.48 | 34.49 | 0.81 | 3.40 |
| Stratified by forest types | 15.29 | 10.88 | 37.94 | 26.99 | 0.90 | 24.40 |
| Unstratified | 13.69 | 9.74 | 50.19 | 35.70 | 0.79 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Ren, C.; Zhang, B.; Wang, Z.; Man, W.; Liu, M. Improved Object-Based Mapping of Aboveground Biomass Using Geographic Stratification with GEDI Data and Multi-Sensor Imagery. Remote Sens. 2023, 15, 2625. https://doi.org/10.3390/rs15102625
Chen L, Ren C, Zhang B, Wang Z, Man W, Liu M. Improved Object-Based Mapping of Aboveground Biomass Using Geographic Stratification with GEDI Data and Multi-Sensor Imagery. Remote Sensing. 2023; 15(10):2625. https://doi.org/10.3390/rs15102625
Chicago/Turabian StyleChen, Lin, Chunying Ren, Bai Zhang, Zongming Wang, Weidong Man, and Mingyue Liu. 2023. "Improved Object-Based Mapping of Aboveground Biomass Using Geographic Stratification with GEDI Data and Multi-Sensor Imagery" Remote Sensing 15, no. 10: 2625. https://doi.org/10.3390/rs15102625