A Discriminative Model for Early Detection of Anthracnose in Strawberry Plants Based on Hyperspectral Imaging Technology
Abstract
:1. Introduction
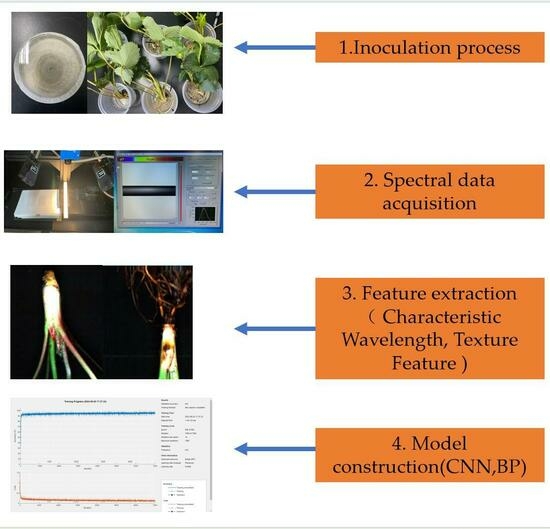
2. Materials and Methods
2.1. Strawberry Plants and Isolates
2.2. Inoculation of Strawberry Anthracnose Pathogen
2.3. Hyperspectral Data Acquisition and Processing
2.4. Methods for Constructing Hyperspectral Models
2.4.1. Spectral Data Extraction
2.4.2. Characteristic Wavelength Selection
2.4.3. MFN Transform and Extraction of Texture Features
2.4.4. Classification Recognition Model Construction
2.4.5. Model Assessment
3. Results
3.1. Changes in Reflectance Caused by Anthracnose
3.2. Characteristic Wavelength Extraction Results and Analysis
3.3. Texture Feature Extraction Results and Analysis
3.4. Recognition Results and Evaluation Based on Different Modeling Methods
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A



References
- Ji, Y.; Li, X.; Gao, Q.; Geng, C.; Duan, K. Colletotrichum species pathogenic to strawberry: Discovery history, global diversity, prevalence in China, and the host range of top two species. Phytopathol. Res. 2022, 4, 42. [Google Scholar] [CrossRef]
- Chen, X.Y.; Dai, D.J.; Zhao, S.F.; Shen, Y.; Wang, H.D.; Zhang, C.Q. Genetic diversity of Colletotrichum spp. causing strawberry anthracnose in Zhejiang, China. Plant Dis. 2020, 104, 1351–1357. [Google Scholar] [CrossRef] [PubMed]
- Jian, Y.; Li, Y.; Tang, G.; Zheng, X.; Khaskheli, M.I.; Gong, G. Identification of Colletotrichum species associated with anthracnose disease of strawberry in Sichuan province, China. Plant Dis. 2021, 105, 3025–3036. [Google Scholar] [CrossRef] [PubMed]
- Soares, V.F.; Velho, A.C.; Carachenski, A.; Astolfi, P.; Stadnik, M.J. First report of Colletotrichum karstii causing anthracnose on strawberry in Brazil. Plant Dis. 2021, 105, 3295. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.J. Epidemiology and pathology of strawberry anthracnose: A North American perspective. HortScience 2008, 43, 69–73. [Google Scholar] [CrossRef]
- Yang, J.; Duan, K.; Liu, Y.; Song, L.; Gao, Q.H. Method to detect and quantify colonization of anthracnose causal agent Colletotrichum gloeosporioides species complex in strawberry by real-time PCR. J. Phytopathol. 2022, 170, 326–336. [Google Scholar] [CrossRef]
- Shengfan, H.; Junjie, L.; Tengfei, X.; Xuefeng, L.; Xiaofeng, L.; Su, L.; Hongqing, W. Simultaneous detection of three crown rot pathogens in field-grown strawberry plants using a multiplex PCR assay. Crop Prot. 2022, 156, 105957. [Google Scholar] [CrossRef]
- Miftakhurohmah; Mutaqin, K.H.; Soekarno, B.P.W.; Wahyuno, D.; Hidayat, S.H. Identification of endogenous and episomal piper yellow mottle virus from the leaves and berries of black pepper (Piper nigrum). Austral. Plant Pathol. 2021, 50, 431–434. [Google Scholar] [CrossRef]
- Schoelz, J.; Volenberg, D.; Adhab, M.; Fang, Z.; Klassen, V.; Spinka, C.; Rwahnih, M.A. A survey of viruses found in grapevine cultivars grown in Missouri. Am. J. Enol. Vitic. 2021, 72, 73–84. [Google Scholar] [CrossRef]
- Khudhair, M.; Obanor, F.; Kazan, K.; Gardiner, D.M.; Aitken, E.; McKay, A.; Giblot-Ducray, D.; Simpfendorfer, S.; Thatcher, L.F. Genetic diversity of Australian Fusarium pseudograminearum populations causing crown rot in wheat. Eur. J. Plant Pathol. 2021, 159, 741–753. [Google Scholar] [CrossRef]
- Lowe, A.; Harrison, N.; French, A.P. Hyperspectral image analysis techniques for the detection and classification of the early onset of plant disease and stress. Plant Methods 2017, 13, 80. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Yang, G.; Pan, Y.; Yang, X.; Chen, L.; Zhao, C. A review of advanced technologies and development for Hyperspectral-based plant disease detection in the past three decades. Remote Sens. 2020, 12, 3188. [Google Scholar] [CrossRef]
- Terentev, A.; Dolzhenko, V.; Fedotov, A.; Eremenko, D. Current state of hyperspectral remote sensing for early plant disease detection: A review. Sensors 2022, 22, 757. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, Z.; Zheng, L.; Han, C.; Wang, X.; Xu, J.; Wang, X. Research progress on the early monitoring of pine wilt disease using hyperspectral techniques. Sensors 2020, 20, 3729. [Google Scholar] [CrossRef] [PubMed]
- Che Ya, N.N.; Mohidem, N.A.; Roslin, N.A.; Saberioon, M.; Tarmidi, M.Z.; Arif Shah, J.; Fazlil Ilahi, W.F.; Man, N. Mobile computing for pest and disease management using spectral signature analysis: A review. Agronomy 2022, 12, 967. [Google Scholar] [CrossRef]
- Yuan, L.; Yan, P.; Han, W.; Huang, Y.; Wang, B.; Zhang, J.; Zhang, H.; Bao, Z. Detection of anthracnose in tea plants based on hyperspectral imaging. Comput. Electron. Agric. 2019, 167, 105039. [Google Scholar] [CrossRef]
- Appeltans, S.; Pieters, J.G.; Mouazen, A.M. Detection of leek white tip disease under field conditions using hyperspectral proximal sensing and supervised machine learning. Comput. Electron. Agric. 2021, 190, 106453. [Google Scholar] [CrossRef]
- Pérez-Roncal, C.; Arazuri, S.; Lopez-Molina, C.; Jarén, C.; Santesteban, L.G.; López-Maestresalas, A. Exploring the potential of hyperspectral imaging to detect Esca disease complex in asymptomatic grapevine leaves. Comput. Electron. Agric. 2022, 196, 106863. [Google Scholar] [CrossRef]
- Xuan, G.; Li, Q.; Shao, Y.; Shi, Y. Early diagnosis and pathogenesis monitoring of wheat powdery mildew caused by Blumeria graminis using hyperspectral imaging. Comput. Electron. Agric. 2022, 197, 106921. [Google Scholar] [CrossRef]
- Xie, C.; Yang, C.; He, Y. Hyperspectral imaging for classification of healthy and gray mold diseased tomato leaves with different infection severities. Comput. Electron. Agric. 2017, 135, 154–162. [Google Scholar] [CrossRef]
- Zhang, J.; Tian, Y.; Yan, L.; Wang, B.; Wang, L.; Xu, J.; Wu, K. Diagnosing the symptoms of sheath blight disease on rice stalk with an in-situ hyperspectral imaging technique. Biosyst. Eng. 2021, 209, 94–105. [Google Scholar] [CrossRef]
- Fazari, A.; Pellicer-Valero, O.J.; Gómez-Sanchıs, J.; Bernardi, B.; Cubero, S.; Benalia, S.; Zimbalatti, G.; Blasco, J. Application of deep convolutional neural networks for the detection of anthracnose in olives using VIS/NIR hyperspectral images. Comput. Electron. Agric. 2021, 187, 106252. [Google Scholar] [CrossRef]
- Gao, Z.; Khot, L.R.; Naidu, R.A.; Zhang, Q. Early detection of grapevine leafroll disease in a red-berried wine grape cultivar using hyperspectral imaging. Comput. Electron. Agric. 2020, 179, 105807. [Google Scholar] [CrossRef]
- Conrad, A.O.; Li, W.; Lee, D.Y.; Wang, G.L.; Rodriguez-Saona, L.; Bonello, P. Machine Learning-based presymptomatic detection of rice sheath blight using -spectral profiles. Plant Phenomics 2020, 2020, 8954085. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Jie, L.; Wang, S.; Qi, H.; Li, S. Classifying wheat hyperspectral pixels of healthy heads and fusarium head blight disease using a deep neural network in the wild field. Remote Sens. 2018, 10, 395. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, J.; Huang, Y.; Tian, Y.; Yuan, L. Detection and discrimination of disease and insect stress of tea plants using hyperspectral imaging combined with wavelet analysis. Comput. Electron. Agric. 2022, 193, 106717. [Google Scholar] [CrossRef]
- Sha, W.; Hu, K.; Weng, S. Statistic and network features of RGB and hyperspectral Imaging for determination of black root mold infection in apples. Foods 2023, 12, 1608. [Google Scholar] [CrossRef] [PubMed]
- Cen, Y.; Huang, Y.; Hu, S.; Zhang, L.; Zhang, J. Early detection of bacterial wilt in tomato with portable hyperspectral spectrometer. Remote Sens. 2022, 14, 2882. [Google Scholar] [CrossRef]
- Jiang, Q.; Wu, G.; Tian, C.; Li, N.; Yang, H.; Bai, Y.; Zhang, B. Hyperspectral imaging for early identification of strawberry leaves diseases with machine learning and spectral fingerprint features. Infrared Phys. Technol. 2021, 118, 103898. [Google Scholar] [CrossRef]
- Miller-Butler, M.A.; Smith, B.J.; Curry, K.J.; Blythe, E.K. Evaluation of detached strawberry leaves for anthracnose disease severity using image analysis and visual ratings. HortScience 2019, 54, 2111–2117. [Google Scholar] [CrossRef]
- Genangeli, A.; Allasia, G.; Bindi, M.; Cantini, C.; Cavaliere, A.; Genesio, L.; Giannotta, G.; Miglietta, F.; Gioli, B. A novel hyperspectral method to detect moldy core in apple fruits. Sensors 2022, 22, 4479. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Zhao, D.; Guan, Q.; Li, J.; Liu, Z.; Jin, Z.; Li, G.; Xu, T. A deep convolutional neural network-based wavelength selection method for spectral characteristics of rice blast disease. Comput. Electron. Agric. 2022, 199, 107199. [Google Scholar] [CrossRef]
- Wu, J.P.; Zhou, J.; Jiao, Z.B.; Fu, J.P.; Xiao, Y.; Guo, F.L. Amorphophallus konjac anthracnose caused by Colletotrichum siamense in China. J. Appl. Microbiol. 2020, 128, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Liu, S.; Yang, X.; Ma, Y.; Zhang, B.; Chu, W. Research on camouflage recognition in simulated operational environment based on hyperspectral imaging technology. J. Spectrosc. 2021, 2021, 6629661. [Google Scholar] [CrossRef]
- Barreto, A.; Paulus, S.; Varrelmann, M.; Mahlein, A. Hyperspectral imaging of symptoms induced by Rhizoctonia solani in sugar beet: Comparison of input data and different machine learning algorithms. J. Plant Dis. Prot. 2020, 127, 441–451. [Google Scholar] [CrossRef]
- Wei, X.; Johnson, M.A.; Langston, D.B.; Mehl, H.L.; Li, S. Identifying optimal wavelengths as disease signatures using hyperspectral sensor and machine learning. Remote Sens. 2021, 13, 2833. [Google Scholar] [CrossRef]
- An, C.; Yan, X.; Lu, C.; Zhu, X. Effect of spectral pretreatment on qualitative identification of adulterated bovine colostrum by near-infrared spectroscopy. Infrared Phys. Technol. 2021, 118, 103869. [Google Scholar] [CrossRef]
- Zhu, H.; Chu, B.; Zhang, C.; Liu, F.; Jiang, L.; He, Y. Hyperspectral imaging for presymptomatic detection of tobacco disease with successive projections algorithm and Machine-learning classifiers. Sci. Rep. 2017, 7, 4125. [Google Scholar] [CrossRef]
- Liu, S.; Yu, H.; Sui, Y.; Zhou, H.; Zhang, J.; Kong, L.; Dang, J.; Zhang, L. Classification of soybean frogeye leaf spot disease using leaf hyperspectral reflectance. PLoS ONE 2021, 16, e257008. [Google Scholar] [CrossRef]
- Liu, N.; Qiao, L.; Xing, Z.; Li, M.; Sun, H.; Zhang, J.; Zhang, Y. Detection of chlorophyll content in growth potato based on spectral variable analysis. Spectr. Lett. 2020, 53, 476–488. [Google Scholar] [CrossRef]
- Zhang, B.; Li, J.; Fan, S.; Huang, W.; Zhao, C.; Liu, C.; Huang, D. Hyperspectral imaging combined with multivariate analysis and band math for detection of common defects on peaches (Prunus persica). Comput. Electron. Agric. 2015, 114, 14–24. [Google Scholar] [CrossRef]
- Liu, L.; Dong, Y.; Huang, W.; Du, X.; Ma, H. Monitoring wheat fusarium head blight using unmanned aerial vehicle hyperspectral imagery. Remote Sens. 2020, 12, 3811. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, Y.; Feng, X.; Xu, H.; Chen, J.; He, Y. Identification of bacterial blight resistant rice seeds using terahertz imaging and hyperspectral imaging combined with convolutional neural network. Front. Plant Sci. 2020, 11, 821. [Google Scholar] [CrossRef] [PubMed]
- Riefolo, C.; Antelmi, I.; Castrignanò, A.; Ruggieri, S.; Galeone, C.; Belmonte, A.; Muolo, M.R.; Ranieri, N.A.; Labarile, R.; Gadaleta, G.; et al. Assessment of the hyperspectral data analysis as a tool to diagnose Xylella fastidiosa in the asymptomatic leaves of olive plants. Plants 2021, 10, 683. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, G.A. Quantifying chlorophylls and caroteniods at leaf and canopy scales: An evaluation of some hyperspectral approaches. Remote Sens. Environ. 1998, 66, 273–285. [Google Scholar] [CrossRef]
- Bruning, B.; Liu, H.; Brien, C.; Berger, B.; Lewis, M.; Garnett, T. The development of hyperspectral distribution maps to predict the content and distribution of nitrogen and water in wheat (Triticum aestivum). Front. Plant Sci. 2019, 10, 1380. [Google Scholar] [CrossRef] [PubMed]
- Eshkabilov, S.; Lee, A.; Sun, X.; Lee, C.W.; Simsek, H. Hyperspectral imaging techniques for rapid detection of nutrient content of hydroponically grown lettuce cultivars. Comput. Electron. Agric. 2021, 181, 105968. [Google Scholar] [CrossRef]
- Wu, G.; Fang, Y.; Jiang, Q.; Cui, M.; Li, N.; Ou, Y.; Diao, Z.; Zhang, B. Early identification of strawberry leaves disease utilizing hyperspectral imaging combing with spectral features, multiple vegetation indices and textural features. Comput. Electron. Agric. 2023, 204, 107553. [Google Scholar] [CrossRef]
- Khairunniza-Bejo, S.; Shahibullah, M.S.; Azmi, A.N.N.; Jahari, M. Non-destructive detection of asymptomatic Ganoderma boninense infection of oil palm seedlings using NIR-hyperspectral data and support vector machine. Appl. Sci. 2021, 11, 10878. [Google Scholar] [CrossRef]
- Pane, C.; Manganiello, G.; Nicastro, N.; Cardi, T.; Carotenuto, F. Powdery mildew caused by Erysiphe cruciferarum on wild rocket (Diplotaxis tenuifolia): Hyperspectral imaging and machine learning modeling for non-destructive disease detection. Agriculture 2021, 11, 337. [Google Scholar] [CrossRef]
- Zhao, J.; Fang, Y.; Chu, G.; Yan, H.; Hu, L.; Huang, L. Identification of Leaf-scale wheat powdery mildew (Blumeria graminis f. sp. tritici) combining hyperspectral imaging and an SVM classifier. Plants 2020, 9, 936. [Google Scholar] [CrossRef]
- Guo, A.; Huang, W.; Ye, H.; Dong, Y.; Ma, H.; Ren, Y.; Ruan, C. Identification of wheat yellow rust using spectral and texture features of hyperspectral images. Remote Sens. 2020, 12, 1419. [Google Scholar] [CrossRef]












| Algorithm | Number of Characteristic Wavelengths | Characteristic Wavelengths (nm) |
|---|---|---|
| SPA | 5 | 945, 901, 927, 591, 833 |
| CARS | 14 | 561, 564, 566, 583, 585, 591, 600, 602, 628, 647, 719, 749, 751, 857 |
| IRF | 11 | 566, 602, 604, 630, 767, 769, 804, 806, 905, 923, 927 |
| Feature | Number of Feature Selection | Accuracy Rate (%) | Time (s) | |
|---|---|---|---|---|
| Train Set | Test Set | |||
| SPA | 5 | 92.83 | 92.50 | 5.4 |
| CARS | 14 | 93.22 | 93.17 | 8.1 |
| IRF | 11 | 94.83 | 94.67 | 7.0 |
| TF | 12 | 75.33 | 75.50 | 7.2 |
| SPA + TF | 17 | 96.56 | 93.67 | 10.1 |
| CARS + TF | 26 | 95.17 | 93.50 | 11.4 |
| IRF + TF | 23 | 96.17 | 95.83 | 10.9 |
| Feature | Number of Feature Selection | Accuracy Rate (%) | Time (s) | |
|---|---|---|---|---|
| Train Set | Test Set | |||
| SPA | 5 | 94.78 | 94.50 | 95 |
| CARS | 14 | 95.61 | 93.83 | 101 |
| IRF | 11 | 95.61 | 94.50 | 97 |
| TF | 12 | 90 | 79.50 | 99 |
| SPA + TF | 17 | 100 | 95.33 | 110 |
| CARS + TF | 26 | 100 | 95.83 | 118 |
| IRF + TF | 23 | 100 | 95.50 | 114 |
| Model | Time/s | P/% | R/% | F1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| H | AI | SI | H | AI | SI | H | AI | SI | ||
| SPA + BP | 5.4 | 99.00 | 90.50 | 88.50 | 99.50 | 87.40 | 91.10 | 0.9925 | 0.8892 | 0.8978 |
| CARS + BP | 8.1 | 100.00 | 86.60 | 93.50 | 100.00 | 93.30 | 86.90 | 1.0000 | 0.8983 | 0.9008 |
| IRF + BP | 7 | 100.00 | 90.00 | 93.40 | 100.00 | 93.80 | 89.50 | 1.0000 | 0.9186 | 0.9141 |
| TF + BP | 7.2 | 72.30 | 75.00 | 80.10 | 84.80 | 67.50 | 74.70 | 0.7805 | 0.7105 | 0.7731 |
| SPA + TF + BP | 10.1 | 100.00 | 90.50 | 90.30 | 99.50 | 91.40 | 89.80 | 0.9975 | 0.9095 | 0.9005 |
| CARS + TF + BP | 11.4 | 99.50 | 90.90 | 90.70 | 99.50 | 91.30 | 90.20 | 0.9950 | 0.9110 | 0.9045 |
| IRF + TF + BP | 10.9 | 99.10 | 93.50 | 94.50 | 100.00 | 94.00 | 93.00 | 0.9955 | 0.9375 | 0.9374 |
| SPA + CNN | 95 | 99.00 | 89.80 | 94.80 | 100.00 | 93.20 | 90.90 | 0.9950 | 0.9147 | 0.9281 |
| CARS + CNN | 101 | 100.00 | 90.70 | 91.00 | 100.00 | 91.10 | 90.50 | 1.0000 | 0.9090 | 0.9075 |
| IRF + CNN | 97 | 100.00 | 89.60 | 93.80 | 100.00 | 94.50 | 88.40 | 1.0000 | 0.9198 | 0.9102 |
| TF + CNN | 99 | 78.90 | 76.80 | 82.40 | 80.50 | 69.80 | 88.30 | 0.7969 | 0.7313 | 0.8525 |
| SPA + TF + CNN | 110 | 96.50 | 93.90 | 95.60 | 99.00 | 92.50 | 94.60 | 0.9773 | 0.9319 | 0.9510 |
| CARS + TF + CNN | 118 | 98.50 | 93.10 | 95.90 | 99.50 | 94.50 | 93.50 | 0.9900 | 0.9380 | 0.9468 |
| IRF + TF + CNN | 114 | 98.00 | 96.50 | 92.10 | 99.00 | 91.10 | 96.90 | 0.9850 | 0.9372 | 0.9444 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Cao, Y.; Wu, E.; Yang, R.; Xu, H.; Qiao, Y. A Discriminative Model for Early Detection of Anthracnose in Strawberry Plants Based on Hyperspectral Imaging Technology. Remote Sens. 2023, 15, 4640. https://doi.org/10.3390/rs15184640
Liu C, Cao Y, Wu E, Yang R, Xu H, Qiao Y. A Discriminative Model for Early Detection of Anthracnose in Strawberry Plants Based on Hyperspectral Imaging Technology. Remote Sensing. 2023; 15(18):4640. https://doi.org/10.3390/rs15184640
Chicago/Turabian StyleLiu, Chao, Yifei Cao, Ejiao Wu, Risheng Yang, Huanliang Xu, and Yushan Qiao. 2023. "A Discriminative Model for Early Detection of Anthracnose in Strawberry Plants Based on Hyperspectral Imaging Technology" Remote Sensing 15, no. 18: 4640. https://doi.org/10.3390/rs15184640