Self-Adaptive-Filling Deep Convolutional Neural Network Classification Method for Mountain Vegetation Type Based on High Spatial Resolution Aerial Images
Abstract
:1. Introduction
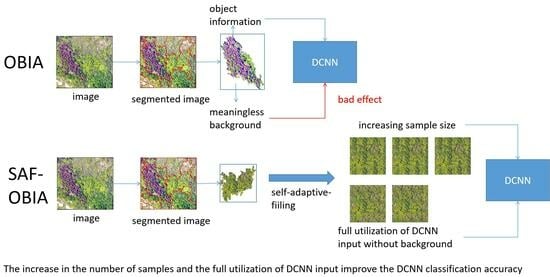
- We propose a self-adaptive-filling based OBIA to enhance the classification performance of a DCNN classifier using the OBIA technique (SAF-DCNN) for the remote sensing images of mountain vegetation.
- To demonstrate the advantages of SAF-based OBIA, we also compare the performance of using DCNN and random forest classifiers under SAF-based OBIA and traditional OBIA at two different data volumes.
- In addition, we compare the performance of OBIA-based methods with semantic segmentation methods on remote sensing images of mountain vegetation by comparing commonly used methods for the semantic segmentation of remote sensing images.
2. Materials and Methods
2.1. Study Area
2.2. Data
2.2.1. Data and Preprocessing
2.2.2. Sample Data for Training and Verification
2.3. Study Methods
2.3.1. Traditional Based OBIA
2.3.2. Self-Adaptive-Filling Based OBIA
2.3.3. Filled Image Generation for SAF-Based OBIA
| Algorithm 1: Self-adaptive-filling Algorithm |
Data: An image A of size Result: An array of n images of size
|
2.3.4. RF and DCNN Using SAF-Based OBIA for Experiments
| Feature Category | Feature Name | Calculation Formula | Description |
|---|---|---|---|
| Spectral characteristics | Mean | Grayscale values of all image elements in the kth band | |
| Standard Deviation | Standard deviation of grayscale values of all image elements in the kth band | ||
| Ratio | The ratio of the mean grayscale value of the image in the kth band to the overall brightness of the object | ||
| Texture characteristics | Entropy | Reflects the amount of information in the image object | |
| Homogeneity | Reflects the intrinsic variability of the image object and the smaller the variance the larger the value | ||
| Contrast | Reflects the degree of change in image objects and highlights anomalies | ||
| Correlation | Reflects the degree of linear correlation of grayscale within the image object | ||
| Angle Second Moment | Reflects the uniformity of grayscale distribution within the image object | ||
| Mean | Reflects the average grayscale within the image object | ||
| Standard Deviation | Reflects the magnitude of grayscale changes within the image object | ||
| Dissimilarity | Reflects the degree of grayscale detail variation within the image object | ||
| Vegetation Index | EXG | Over Green Index | |
| EXR | Super Red Index | ||
| EXGR | Super Green Super Red Differential Index | ||
| NGBD I | Normalized Green and Blue Disparity Index | ||
| RGBV I | Red, Green, and Blue Vegetation Index | ||
| NGRD I | Normalized Red–Green Variance Index |
2.3.5. RF and DCNN Using OBIA for Comparison
2.3.6. Semantic Segmentation: U-net, MACU-net, and SegNeXt for Comparison
2.3.7. Experiment Design
3. Results
3.1. Overall Accuracy Evaluation
3.2. Accuracy of Each Vegetation
3.3. Classification Results Map
4. Discussion
4.1. Performance of SAF-DCNN on Mountain Vegetation Classification
4.2. Performance of SAF-Based OBIA
4.3. Comparison of OBIA and Semantic Segmentation in Mountain Vegetation Classification
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pettorelli, N.; Schulte to Bühne, H.; Tulloch, A.; Dubois, G.; Macinnis-Ng, C.; Queirós, A.M.; Keith, D.A.; Wegmann, M.; Schrodt, F.; Stellmes, M.; et al. Satellite remote sensing of ecosystem functions: Opportunities, challenges and way forward. Remote Sens. Ecol. Conserv. 2018, 4, 71–93. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote sensing technologies for enhancing forest inventories: A review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef]
- Jurado, J.M.; López, A.; Pádua, L.; Sousa, J.J. Remote sensing image fusion on 3D scenarios: A review of applications for agriculture and forestry. Int. J. Appl. Earth Obs. Geoinf. 2022, 112, 102856. [Google Scholar] [CrossRef]
- Atzberger, C.; Darvishzadeh, R.; Schlerf, M.; Le Maire, G. Suitability and adaptation of PROSAIL radiative transfer model for hyperspectral grassland studies. Remote Sens. Lett. 2013, 4, 55–64. [Google Scholar] [CrossRef]
- Mulla, D.J. Twenty five years of remote sensing in precision agriculture: Key advances and remaining knowledge gaps. Biosyst. Eng. 2013, 114, 358–371. [Google Scholar] [CrossRef]
- Becker, A.; Russo, S.; Puliti, S.; Lang, N.; Schindler, K.; Wegner, J.D. Country-wide retrieval of forest structure from optical and SAR satellite imagery with deep ensembles. ISPRS J. Photogramm. Remote Sens. 2023, 195, 269–286. [Google Scholar] [CrossRef]
- Jamison, E.A.K.; D’Amato, A.W.; Dodds, K.J. Describing a landscape mosaic: Forest structure and composition across community types and management regimes in inland northeastern pitch pine barrens. For. Ecol. Manag. 2023, 536, 120859. [Google Scholar] [CrossRef]
- Rybansky, M. Determination of Forest Structure from Remote Sensing Data for Modeling the Navigation of Rescue Vehicles. Appl. Sci. 2022, 12, 3939. [Google Scholar] [CrossRef]
- Ranjan, R. Linking green bond yields to the species composition of forests for improving forest quality and sustainability. J. Clean. Prod. 2022, 379, 134708. [Google Scholar] [CrossRef]
- Edelmann, P.; Ambarlı, D.; Gossner, M.M.; Schall, P.; Ammer, C.; Wende, B.; Schulze, E.D.; Weisser, W.W.; Seibold, S. Forest management affects saproxylic beetles through tree species composition and canopy cover. For. Ecol. Manag. 2022, 524, 120532. [Google Scholar] [CrossRef]
- Nasiri, V.; Beloiu, M.; Darvishsefat, A.A.; Griess, V.C.; Maftei, C.; Waser, L.T. Mapping tree species composition in a Caspian temperate mixed forest based on spectral-temporal metrics and machine learning. Int. J. Appl. Earth Obs. Geoinf. 2023, 116, 103154. [Google Scholar] [CrossRef]
- Cavender-Bares, J.; Schneider, F.D.; Santos, M.J.; Armstrong, A.; Carnaval, A.; Dahlin, K.M.; Fatoyinbo, L.; Hurtt, G.C.; Schimel, D.; Townsend, P.A.; et al. Integrating remote sensing with ecology and evolution to advance biodiversity conservation. Nat. Ecol. Evol. 2022, 6, 506–519. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Avtar, R.; Umarhadi, D.A.; Louw, A.S.; Shrivastava, S.; Yunus, A.P.; Khedher, K.M.; Takemi, T.; Shibata, H. Post-typhoon forest damage estimation using multiple vegetation indices and machine learning models. Weather Clim. Extrem. 2022, 38, 100494. [Google Scholar] [CrossRef]
- Peereman, J.; Hogan, J.A.; Lin, T.C. Intraseasonal interactive effects of successive typhoons characterize canopy damage of forests in Taiwan: A remote sensing-based assessment. For. Ecol. Manag. 2022, 521, 120430. [Google Scholar] [CrossRef]
- Pawlik, Ł.; Harrison, S.P. Modelling and prediction of wind damage in forest ecosystems of the Sudety Mountains, SW Poland. Sci. Total Environ. 2022, 815, 151972. [Google Scholar] [CrossRef]
- Marlier, M.E.; Resetar, S.A.; Lachman, B.E.; Anania, K.; Adams, K. Remote sensing for natural disaster recovery: Lessons learned from Hurricanes Irma and Maria in Puerto Rico. Environ. Sci. Policy 2022, 132, 153–159. [Google Scholar] [CrossRef]
- Sun, Y.; Huang, J.; Ao, Z.; Lao, D.; Xin, Q. Deep learning approaches for the mapping of tree species diversity in a tropical wetland using airborne LiDAR and high-spatial-resolution remote sensing images. Forests 2019, 10, 1047. [Google Scholar] [CrossRef]
- Sothe, C.; De Almeida, C.; Schimalski, M.B.; Liesenberg, V.; La Rosa, L.; Castro, J.; Feitosa, R.Q. A comparison of machine and deep-learning algorithms applied to multisource data for a subtropical forest area classification. Int. J. Remote Sens. 2020, 41, 1943–1969. [Google Scholar] [CrossRef]
- Wagner, F.H.; Sanchez, A.; Tarabalka, Y.; Lotte, R.G.; Ferreira, M.P.; Aidar, M.P.; Gloor, E.; Phillips, O.L.; Aragao, L.E. Using the U-net convolutional network to map forest types and disturbance in the Atlantic rainforest with very high resolution images. Remote Sens. Ecol. Conserv. 2019, 5, 360–375. [Google Scholar] [CrossRef]
- Kattenborn, T.; Eichel, J.; Fassnacht, F.E. Convolutional Neural Networks enable efficient, accurate and fine-grained segmentation of plant species and communities from high-resolution UAV imagery. Sci. Rep. 2019, 9, 17656. [Google Scholar] [CrossRef] [PubMed]
- Fricker, G.A.; Ventura, J.D.; Wolf, J.A.; North, M.P.; Davis, F.W.; Franklin, J. A convolutional neural network classifier identifies tree species in mixed-conifer forest from hyperspectral imagery. Remote Sens. 2019, 11, 2326. [Google Scholar] [CrossRef]
- Jiang, S.; Yao, W.; Heurich, M. Dead wood detection based on semantic segmentation of VHR aerial CIR imagery using optimized FCN-Densenet. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, 42, 127–133. [Google Scholar] [CrossRef]
- Ye, W.; Lao, J.; Liu, Y.; Chang, C.C.; Zhang, Z.; Li, H.; Zhou, H. Pine pest detection using remote sensing satellite images combined with a multi-scale attention-UNet model. Ecol. Inform. 2022, 72, 101906. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, Y.; Wang, M.; Shen, Q.; Huang, J. Object-scale adaptive convolutional neural networks for high-spatial resolution remote sensing image classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 14, 283–299. [Google Scholar] [CrossRef]
- Shui, W.; Li, H.; Zhang, Y.; Jiang, C.; Zhu, S.; Wang, Q.; Liu, Y.; Zong, S.; Huang, Y.; Ma, M. Is an Unmanned Aerial Vehicle (UAV) Suitable for Extracting the Stand Parameters of Inaccessible Underground Forests of Karst Tiankeng? Remote Sens. 2022, 14, 4128. [Google Scholar] [CrossRef]
- Zhu, Y.; Zeng, Y.; Zhang, M. Extract of land use/cover information based on HJ satellites data and object-oriented classification. Trans. Chin. Soc. Agric. Eng. 2017, 33, 258–265. [Google Scholar]
- Chen, H.; Yin, D.; Chen, J.; Chen, Y. Automatic Spectral Representation With Improved Stacked Spectral Feature Space Patch (ISSFSP) for CNN-Based Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2022, 60, 4709014. [Google Scholar] [CrossRef]
- Sidike, P.; Sagan, V.; Maimaitijiang, M.; Maimaitiyiming, M.; Shakoor, N.; Burken, J.; Mockler, T.; Fritschi, F.B. dPEN: Deep Progressively Expanded Network for mapping heterogeneous agricultural landscape using WorldView-3 satellite imagery. Remote Sens. Environ. 2019, 221, 756–772. [Google Scholar] [CrossRef]
- Zhang, C.; Pan, X.; Li, H.; Gardiner, A.; Sargent, I.; Hare, J.; Atkinson, P.M. A hybrid MLP-CNN classifier for very fine resolution remotely sensed image classification. ISPRS J. Photogramm. Remote Sens. 2018, 140, 133–144. [Google Scholar] [CrossRef]
- Liu, T.; Abd-Elrahman, A.; Morton, J.; Wilhelm, V.L. Comparing fully convolutional networks, random forest, support vector machine, and patch-based deep convolutional neural networks for object-based wetland mapping using images from small unmanned aircraft system. GIScience Remote Sens. 2018, 55, 243–264. [Google Scholar] [CrossRef]
- Ma, L.; Li, M.; Ma, X.; Cheng, L.; Du, P.; Liu, Y. A review of supervised object-based land-cover image classification. ISPRS J. Photogramm. Remote Sens. 2017, 130, 277–293. [Google Scholar] [CrossRef]
- dos Santos Ferreira, A.; Freitas, D.M.; da Silva, G.G.; Pistori, H.; Folhes, M.T. Weed detection in soybean crops using ConvNets. Comput. Electron. Agric. 2017, 143, 314–324. [Google Scholar] [CrossRef]
- Hartling, S.; Sagan, V.; Sidike, P.; Maimaitijiang, M.; Carron, J. Urban tree species classification using a WorldView-2/3 and LiDAR data fusion approach and deep learning. Sensors 2019, 19, 1284. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Abd-Elrahman, A. Deep convolutional neural network training enrichment using multi-view object-based analysis of Unmanned Aerial systems imagery for wetlands classification. ISPRS J. Photogramm. Remote Sens. 2018, 139, 154–170. [Google Scholar] [CrossRef]
- Office of the Leading Group of the First National Geographic Census of the State Council. Technical Regulations for the Production of Digital Orthophotos; Technical Report 05-2013; Office of the Leading Group of the First National Geographic Census of the State Council: Beijing, China, 2013. [Google Scholar]
- Cheng, G.; Han, J. A survey on object detection in optical remote sensing images. ISPRS J. Photogramm. Remote Sens. 2016, 117, 11–28. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Lobo Torres, D.; Queiroz Feitosa, R.; Nigri Happ, P.; Elena Cué La Rosa, L.; Marcato Junior, J.; Martins, J.; Olã Bressan, P.; Gonçalves, W.N.; Liesenberg, V. Applying fully convolutional architectures for semantic segmentation of a single tree species in urban environment on high resolution UAV optical imagery. Sensors 2020, 20, 563. [Google Scholar] [CrossRef]
- Li, R.; Duan, C.; Zheng, S. Macu-net semantic segmentation from high-resolution remote sensing images. arXiv 2020, arXiv:2007.13083. [Google Scholar]
- Guo, M.H.; Lu, C.Z.; Hou, Q.; Liu, Z.; Cheng, M.M.; Hu, S.m. SegNeXt: Rethinking Convolutional Attention Design for Semantic Segmentation. In Advances in Neural Information Processing Systems; Koyejo, S., Mohamed, S., Agarwal, A., Belgrave, D., Cho, K., Oh, A., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2022; Volume 35, pp. 1140–1156. [Google Scholar]
- Fraiwan, M.; Faouri, E. On the automatic detection and classification of skin cancer using deep transfer learning. Sensors 2022, 22, 4963. [Google Scholar] [CrossRef]
- Fei, H.; Fan, Z.; Wang, C.; Zhang, N.; Wang, T.; Chen, R.; Bai, T. Cotton classification method at the county scale based on multi-features and random forest feature selection algorithm and classifier. Remote Sens. 2022, 14, 829. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, W.; Wei, P.; Fang, P.; Zhang, X.; Yan, N.; Liu, W.; Zhao, H.; Wu, Q. Classification of Zambian grasslands using random forest feature importance selection during the optimal phenological period. Ecol. Indic. 2022, 135, 108529. [Google Scholar] [CrossRef]
- Feizizadeh, B.; Darabi, S.; Blaschke, T.; Lakes, T. QADI as a new method and alternative to kappa for accuracy assessment of remote sensing-based image classification. Sensors 2022, 22, 4506. [Google Scholar] [CrossRef] [PubMed]
- Neyns, R.; Canters, F. Mapping of urban vegetation with high-resolution remote sensing: A review. Remote Sens. 2022, 14, 1031. [Google Scholar] [CrossRef]
- Faruque, M.J.; Vekerdy, Z.; Hasan, M.Y.; Islam, K.Z.; Young, B.; Ahmed, M.T.; Monir, M.U.; Shovon, S.M.; Kakon, J.F.; Kundu, P. Monitoring of land use and land cover changes by using remote sensing and GIS techniques at human-induced mangrove forests areas in Bangladesh. Remote Sens. Appl. Soc. Environ. 2022, 25, 100699. [Google Scholar] [CrossRef]














| Vegetation Type | Picture Taken by UAV | Vegetation Type | Picture Taken by UAV |
|---|---|---|---|
| Phyllostachys Spectabilis |  | Quercus acutissima |  |
| Cunninghamia lanceolate |  | Pinus thunbergii |  |
| Vegetation Type | Image | Description |
|---|---|---|
| Castanea mollissima |  | This is a tree with edible fruits. The leaves of the tree are characterized by their broad shape and haphazard arrangement. They exhibit vibrant hues and distinct spectral features. |
| Quercus acutissima |  | In the study area, Quercus acutissima is widely distributed in the mountain forests due to its remarkable adaptability. Quercus acutissima has smaller leaves than C. mollissima quercus, with dense, naturally occurring branches. Quercus acutissima has an inconspicuous crown, giving a distinctly “broken” silhouette. |
| Pinus thunbergii |  | This tree has a small canopy, little shading, low numbers, and random distribution in this study area. It appears black on the image, rarely pure forest, and is often mixed with green shrubs. |
| Cunninghamia lanceolate |  | This is similar in image to the textural features of the pine but the color differs from those of the P.thunbergii, which is a familiar brown color. It is more dense and slightly taller than the P.thunbergii in the study area. |
| Pinus taeda |  | The color of this vegetation on the image is similar to that of Cunninghamia lanceolate, but the textural features are different and the canopy is more pronounced than in Cunninghamia lanceolate. |
| Camellia sinensis |  | Artificially reclaimed Camellia sinensis with neatly shaped and distinctive features. |
| Phyllostachys Spectabilis |  | This vegetation looks smoother and more finely textured on the image, with distinctive features, but it is sometimes intermixed with trees whose canopies can partially obscure it. |
| Broadleaf Forest |  | A mixture of different species of broadleaf woods. |
| Shrub and Grass |  | This one appears to have no visible canopy on the image, and sometimes contains bare rocks or some other trees. |
| Mixed Broadleaf–Conifer Forest |  | Contains broadleaf and coniferous forests, mixed together. |
| Castanea mollissima | Quercus acutissima | Pinus thunbergii | Cunninghamia lanceolate | Pinus taeda | Camellia sinensis | Phyllostachys Spectabilis | Broadleaf Forest | Shrub and Grass | Mixed Broadleaf–Conifer Forest | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SAF-RF-DR * | PA | 0.177 | 0.565 | 0.547 | 0.520 | 0.546 | 0.800 | 1.000 | 0.354 | 0.647 | 0.467 |
| UA | 0.214 | 0.718 | 0.265 | 0.520 | 0.300 | 0.348 | 0.125 | 0.193 | 0.734 | 0.525 | |
| SAF-DCNN-DR | PA | 0.568 | 0.745 | 0.667 | 0.633 | 0.750 | 0.696 | 0.750 | 0.467 | 0.745 | 0.660 |
| UA | 0.595 | 0.708 | 0.623 | 0.740 | 0.545 | 0.696 | 0.375 | 0.523 | 0.784 | 0.635 | |
| OBIA-RF-DR | PA | 0.286 | 0.463 | 0.497 | 0.471 | 0.500 | 0.667 | 0.500 | 0.300 | 0.644 | 0.401 |
| UA | 0.098 | 0.634 | 0.307 | 0.519 | 0.045 | 0.087 | 0.125 | 0.199 | 0.703 | 0.411 | |
| OBIA-DCNN-DR | PA | 0.632 | 0.814 | 0.628 | 0.652 | 0.069 | 0.875 | 1.000 | 0.473 | 0.724 | 0.719 |
| UA | 0.571 | 0.759 | 0.689 | 0.779 | 0.273 | 0.913 | 0.500 | 0.464 | 0.784 | 0.462 | |
| U-net-DR | PA | 0.403 | 0.596 | 0.460 | 0.536 | 0.342 | 0.611 | 0.451 | 0.255 | 0.603 | 0.488 |
| UA | 0.372 | 0.684 | 0.324 | 0.449 | 0.603 | 0.453 | 0.070 | 0.228 | 0.674 | 0.456 | |
| MACU-net-DR | PA | 0.604 | 0.771 | 0.679 | 0.587 | 0.503 | 0.712 | 0.000 | 0.511 | 0.725 | 0.611 |
| UA | 0.502 | 0.764 | 0.671 | 0.567 | 0.470 | 0.661 | 0.000 | 0.362 | 0.770 | 0.718 | |
| SAF-RF-DP * | PA | 0.750 | 0.477 | 0.478 | 0.432 | 0.250 | 0.556 | 0.000 | 0.274 | 0.632 | 0.409 |
| UA | 0.071 | 0.706 | 0.214 | 0.416 | 0.050 | 0.217 | 0.000 | 0.133 | 0.713 | 0.442 | |
| SAF-DCNN-DP | PA | 0.575 | 0.659 | 0.656 | 0.592 | 0.563 | 1.000 | 0.000 | 0.453 | 0.660 | 0.542 |
| UA | 0.548 | 0.704 | 0.483 | 0.623 | 0.409 | 0.652 | 0.000 | 0.351 | 0.796 | 0.522 | |
| OBIA-RF-DP | PA | 0.000 | 0.432 | 0.450 | 0.421 | 0.000 | 0.000 | 0.000 | 0.234 | 0.577 | 0.292 |
| UA | 0.000 | 0.597 | 0.249 | 0.312 | 0.000 | 0.000 | 0.000 | 0.166 | 0.680 | 0.324 | |
| OBIA-DCNN-DP | PA | 0.190 | 0.670 | 0.571 | 0.613 | 0.800 | 1.000 | 0.000 | 0.364 | 0.635 | 0.526 |
| UA | 0.095 | 0.699 | 0.420 | 0.597 | 0.182 | 0.522 | 0.000 | 0.318 | 0.826 | 0.482 | |
| U-net-DP | PA | 0.014 | 0.261 | 0.149 | 0.060 | 0.010 | 0.031 | 0.010 | 0.076 | 0.277 | 0.105 |
| UA | 0.009 | 0.113 | 0.124 | 0.252 | 0.066 | 0.078 | 0.006 | 0.080 | 0.284 | 0.032 | |
| MACU-net-DP | PA | 0.426 | 0.679 | 0.542 | 0.624 | 0.312 | 0.000 | 0.000 | 0.486 | 0.694 | 0.495 |
| UA | 0.431 | 0.676 | 0.578 | 0.554 | 0.219 | 0.000 | 0.000 | 0.352 | 0.704 | 0.657 |
| Classifiers | SAF-DCNN | OBIA-DCNN |
|---|---|---|
| Fold #1 | 0.662 | 0.635 |
| Fold #2 | 0.651 | 0.621 |
| Fold #3 | 0.649 | 0.652 |
| Fold #4 | 0.729 | 0.688 |
| Fold #5 | 0.694 | 0.658 |
| Fold #6 | 0.698 | 0.653 |
| Fold #7 | 0.686 | 0.667 |
| Fold #8 | 0.675 | 0.630 |
| Fold #9 | 0.674 | 0.649 |
| Fold #10 | 0.673 | 0.665 |
| Mean | 0.679 | 0.651 |
| Pairwise t-test | t:5.549 p:0.00357 1 |
| Region | OA | Kappa | mIoU |
|---|---|---|---|
| Region A | 0.694 | 0.573 | 0.403 |
| Region B | 0.624 | 0.461 | 0.426 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Fei, X.; Chen, P.; Wang, Z.; Gao, Y.; Cheng, K.; Wang, H.; Zhang, Y. Self-Adaptive-Filling Deep Convolutional Neural Network Classification Method for Mountain Vegetation Type Based on High Spatial Resolution Aerial Images. Remote Sens. 2024, 16, 31. https://doi.org/10.3390/rs16010031
Li S, Fei X, Chen P, Wang Z, Gao Y, Cheng K, Wang H, Zhang Y. Self-Adaptive-Filling Deep Convolutional Neural Network Classification Method for Mountain Vegetation Type Based on High Spatial Resolution Aerial Images. Remote Sensing. 2024; 16(1):31. https://doi.org/10.3390/rs16010031
Chicago/Turabian StyleLi, Shiou, Xianyun Fei, Peilong Chen, Zhen Wang, Yajun Gao, Kai Cheng, Huilong Wang, and Yuanzhi Zhang. 2024. "Self-Adaptive-Filling Deep Convolutional Neural Network Classification Method for Mountain Vegetation Type Based on High Spatial Resolution Aerial Images" Remote Sensing 16, no. 1: 31. https://doi.org/10.3390/rs16010031