Extracting Shrubland in Deserts from Medium-Resolution Remote-Sensing Data at Large Scale
Abstract
:1. Introduction
- (1)
- Desert shrubs are relatively sparse and have low aggregation, so they are very difficult to be identified through medium-resolution remote-sensing imagery and even high-resolution remote-sensing imagery;
- (2)
- The areas of shrublands within deserts are very small, so very few samples have been collected while mapping the global land cover by using machine-learning methods, and the samples are too few to learn the characteristics of the shrubland in deserts;
- (3)
- Although shrub is vegetation, and tools such as the higher vegetation index will show the features of vegetation, the dry conditions in the desert usually depress these features; the input data for global land-cover mapping usually cannot cover the key date of vegetation variation.
2. Study Area and Materials
2.1. Study Area
2.2. Remote-Sensing Data and the Land-Cover Datasets
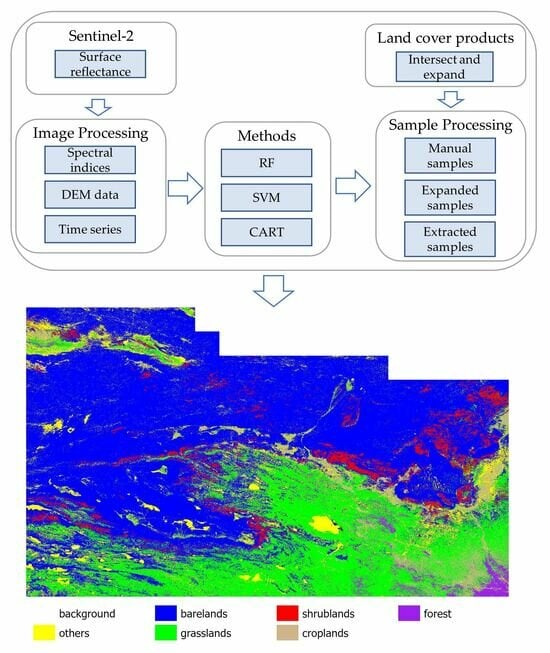
3. Methods
3.1. Feature Construct
3.2. Machine-Learning Modules
3.3. Accuracy Assessment
3.4. Training Data-Retrieving Strategy
3.5. Time Series Composite
4. Results
4.1. Influence of the Feature Variables on the Classification Accuracy
4.2. Influence of the Times-Series Data on the Classification Accuracy
4.3. Classification Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A



| Name | Land Cover Types | OT | BL | GL | SL | PA |
|---|---|---|---|---|---|---|
| GLC-FCS | OT | 293 | 1 | 30 | 0 | 0.904 |
| BL | 205 | 779 | 100 | 20 | 0.706 | |
| GL | 20 | 0 | 376 | 0 | 0.95 | |
| SL | 43 | 172 | 17 | 11 | 0.005 | |
| UA | 0.522 | 0.818 | 0.719 | 0.355 | ||
| OA | 0.706 | |||||
| FROM-GLC | OT | 243 | 34 | 47 | 0 | 0.75 |
| BL | 6 | 1084 | 14 | 0 | 0.982 | |
| GL | 16 | 17 | 361 | 2 | 0.912 | |
| SL | 1 | 238 | 3 | 1 | 0.004 | |
| UA | 0.914 | 0.789 | 0.849 | 0.333 | ||
| OA | 0.817 | |||||
| GlobeLand30 | OT | 286 | 8 | 29 | 1 | 0.883 |
| BL | 11 | 944 | 146 | 3 | 0.855 | |
| GL | 34 | 8 | 349 | 5 | 0.881 | |
| SL | 1 | 206 | 36 | 0 | 0 | |
| UA | 0.861 | 0.81 | 0.623 | 0 | ||
| OA | 0.764 | |||||
| ESA World Cover | OT | 274 | 22 | 24 | 4 | 0.846 |
| BL | 5 | 1062 | 37 | 0 | 0.962 | |
| GL | 20 | 2 | 374 | 0 | 0.944 | |
| SL | 0 | 235 | 6 | 2 | 0.008 | |
| UA | 0.916 | 0.804 | 0.848 | 0.333 | ||
| OA | 0.828 |
References
- Sun, Q.; Zhang, P.; Wei, H.; Liu, A.; You, S.; Sun, D. Improved Mapping and Understanding of Desert Vegetation-Habitat Complexes from Intraannual Series of Spectral Endmember Space Using Cross-Wavelet Transform and Logistic Regression. Remote Sens. Environ. 2020, 236, 111516. [Google Scholar] [CrossRef]
- Yao, Y.; Zhao, Z.; Wei, X.; Shao, M. Effects of Shrub Species on Soil Nitrogen Mineralization in the Desert-Loess Transition Zone. Catena 2019, 173, 330–338. [Google Scholar] [CrossRef]
- Rogan, J.; Chen, D. Remote Sensing Technology for Mapping and Monitoring Land-Cover and Land-Use Change. Prog. Plan. 2004, 61, 301–325. [Google Scholar] [CrossRef]
- Brandt, M.; Hiernaux, P.; Tagesson, T.; Verger, A.; Rasmussen, K.; Diouf, A.A.; Mbow, C.; Mougin, E.; Fensholt, R. Woody Plant Cover Estimation in Drylands from Earth Observation Based Seasonal Metrics. Remote Sens. Environ. 2016, 172, 28–38. [Google Scholar] [CrossRef]
- Cao, X.; Liu, Y.; Liu, Q.; Cui, X.; Chen, X.; Chen, J. Estimating the Age and Population Structure of Encroaching Shrubs in Arid/Semiarid Grasslands Using High Spatial Resolution Remote Sensing Imagery. Remote Sens. Environ. 2018, 216, 572–585. [Google Scholar] [CrossRef]
- Zhou, H.; Fu, L.; Sharma, R.P.; Lei, Y.; Guo, J. A Hybrid Approach of Combining Random Forest with Texture Analysis and VDVI for Desert Vegetation Mapping Based on UAV RGB Data. Remote Sens. 2021, 13, 1891. [Google Scholar] [CrossRef]
- Zhang, T.; Bi, Y.; Du, J.; Zhu, X.; Gao, X. Classification of Desert Grassland Species Based on a Local-Global Feature Enhancement Network and UAV Hyperspectral Remote Sensing. Ecol. Inform. 2022, 72, 101852. [Google Scholar] [CrossRef]
- Mao, P.; Qin, L.; Hao, M.; Zhao, W.; Luo, J.; Qiu, X.; Xu, L.; Xiong, Y.; Ran, Y.; Yan, C.; et al. An Improved Approach to Estimate Above-Ground Volume and Biomass of Desert Shrub Communities Based on UAV RGB Images. Ecol. Indic. 2021, 125, 107494. [Google Scholar] [CrossRef]
- Al-Ali, Z.M.; Abdullah, M.M.; Asadalla, N.B.; Gholoum, M. A Comparative Study of Remote Sensing Classification Methods for Monitoring and Assessing Desert Vegetation Using a UAV-Based Multispectral Sensor. Environ. Monit. Assess. 2020, 192, 389. [Google Scholar] [CrossRef]
- Sun, B.; Li, Z.; Gao, W.; Zhang, Y.; Gao, Z.; Song, Z.; Qin, P.; Tian, X. Identification and Assessment of the Factors Driving Vegetation Degradation/Regeneration in Drylands Using Synthetic High Spatiotemporal Remote Sensing Data—A Case Study in Zhenglanqi, Inner Mongolia, China. Ecol. Indic. 2019, 107, 105614. [Google Scholar] [CrossRef]
- Peng, H.Y.; Li, X.; Tong, S. Effects of Shrub Encroachment on Biomass and Biodiversity in the Typical Steppe of Inner Mongolia. Acta Ecol. Sin. 2013, 33, 7221–7229. [Google Scholar] [CrossRef]
- Laliberte, A.S.; Rango, A.; Havstad, K.M.; Paris, J.F.; Beck, R.F.; McNeely, R.; Gonzalez, A.L. Object-Oriented Image Analysis for Mapping Shrub Encroachment from 1937 to 2003 in Southern New Mexico. Remote Sens. Environ. 2004, 93, 198–210. [Google Scholar] [CrossRef]
- Beck, P.S.A.; Horning, N.; Goetz, S.J.; Loranty, M.M.; Tape, K.D. Shrub Cover on the North Slope of Alaska: A circa 2000 Baseline Map. Arct. Antarct. Alp. Res. 2011, 43, 355–363. [Google Scholar] [CrossRef]
- Baumann, M.; Levers, C.; Macchi, L.; Bluhm, H.; Waske, B.; Gasparri, N.I.; Kuemmerle, T. Mapping Continuous Fields of Tree and Shrub Cover across the Gran Chaco Using Landsat 8 and Sentinel-1 Data. Remote Sens. Environ. 2018, 216, 201–211. [Google Scholar] [CrossRef]
- Bayle, A.; Carlson, B.Z.; Thierion, V.; Isenmann, M.; Choler, P. Improved Mapping of Mountain Shrublands Using the Sentinel-2 Red-Edge Band. Remote Sens. 2019, 11, 2807. [Google Scholar] [CrossRef]
- Vanselow, K.A.; Samimi, C. Predictive Mapping of Dwarf Shrub Vegetation in an Arid High Mountain Ecosystem Using Remote Sensing and Random Forests. Remote Sens. 2014, 6, 6709–6726. [Google Scholar] [CrossRef]
- Gong, P.; Liu, H.; Zhang, M.; Li, C.; Wang, J.; Huang, H.; Clinton, N.; Ji, L.; Li, W.; Bai, Y.; et al. Stable Classification with Limited Sample: Transferring a 30-m Resolution Sample Set Collected in 2015 to Mapping 10-m Resolution Global Land Cover in 2017. Sci. Bull. 2019, 64, 3. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, L.; Chen, X.; Gao, Y.; Xie, S.; Mi, J. GLC_FCS30: Global Land-Cover Product with Fine Classification System at 30 m Using Time-Series Landsat Imagery. Earth Syst. Sci. Data 2021, 13, 2753–2776. [Google Scholar] [CrossRef]
- Chen, J.; Chen, J.; Liao, A.; Cao, X.; Chen, L.; Chen, X.; He, C.; Han, G.; Peng, S.; Lu, M.; et al. Global Land Cover Mapping at 30m Resolution: A POK-Based Operational Approach. ISPRS J. Photogramm. Remote Sens. 2015, 103, 7–27. [Google Scholar] [CrossRef]
- Zanaga, D.; Van De Kerchove, R.; De Keersmaecker, W.; Souverijns, N.; Brockmann, C.; Quast, R.; Wevers, J.; Grosu, A.; Paccini, A.; Vergnaud, S.; et al. ESA WorldCover 10 m 2020 V100 2021. Available online: https://zenodo.org/records/5571936 (accessed on 1 June 2023).
- Drusch, M.; Del Bello, U.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P.; et al. Sentinel-2: ESA’s Optical High-Resolution Mission for GMES Operational Services. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- Huang, S.; Tang, L.; Hupy, J.P.; Wang, Y.; Shao, G. A Commentary Review on the Use of Normalized Difference Vegetation Index (NDVI) in the Era of Popular Remote Sensing. J. For. Res. 2021, 32, 1–6. [Google Scholar] [CrossRef]
- Son, N.T.; Chen, C.F.; Chen, C.R.; Minh, V.Q.; Trung, N.H. A Comparative Analysis of Multitemporal MODIS EVI and NDVI Data for Large-Scale Rice Yield Estimation. Agric. For. Meteorol. 2014, 197, 52–64. [Google Scholar] [CrossRef]
- Gao, B. NDWI—A Normalized Difference Water Index for Remote Sensing of Vegetation Liquid Water from Space. Remote Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Rasul, A.; Balzter, H.; Ibrahim, G.R.F.; Hameed, H.M.; Wheeler, J.; Adamu, B.; Ibrahim, S.; Najmaddin, P.M. Applying Built-Up and Bare-Soil Indices from Landsat 8 to Cities in Dry Climates. Land 2018, 7, 81. [Google Scholar] [CrossRef]
- Zhang, Y.; Odeh, I.O.A.; Han, C. Bi-Temporal Characterization of Land Surface Temperature in Relation to Impervious Surface Area, NDVI and NDBI, Using a Sub-Pixel Image Analysis. Int. J. Appl. Earth Obs. Geoinf. 2009, 11, 256–264. [Google Scholar] [CrossRef]
- Gonenc, A.; Ozerdem, M.S.; Acar, E. Comparison of NDVI and RVI Vegetation Indices Using Satellite Images. In Proceedings of the 2019 8th International Conference on Agro-Geoinformatics (Agro-Geoinformatics), Istanbul, Turkey, 16–19 July 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–4. [Google Scholar]
- Parmar, A.; Katariya, R.; Patel, V. A Review on Random Forest: An Ensemble Classifier. In International Conference on Intelligent Data Communication Technologies and Internet of Things (ICICI) 2018; Hemanth, J., Fernando, X., Lafata, P., Baig, Z., Eds.; Lecture Notes on Data Engineering and Communications Technologies; Springer International Publishing: Cham, Switzerland, 2019; Volume 26, pp. 758–763. ISBN 978-3-030-03145-9. [Google Scholar]
- Belgiu, M.; Drăguţ, L. Random Forest in Remote Sensing: A Review of Applications and Future Directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Shao, Y.; Lunetta, R.S. Comparison of Support Vector Machine, Neural Network, and CART Algorithms for the Land-Cover Classification Using Limited Training Data Points. ISPRS J. Photogramm. Remote Sens. 2012, 70, 78–87. [Google Scholar] [CrossRef]
- Bittencourt, H.R.; Clarke, R.T. Use of Classification and Regression Trees (CART) to Classify Remotely-Sensed Digital Images. In Proceedings of the IGARSS 2003. 2003 IEEE International Geoscience and Remote Sensing Symposium. Proceedings (IEEE Cat. No.03CH37477), Toulouse, France, 21–25 July 2003; Volume 6, pp. 3751–3753. [Google Scholar]
- Congalton, R.G. A Review of Assessing the Accuracy of Classifications of Remotely Sensed Data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-Scale Geospatial Analysis for Everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Pal, M.; Mather, P.M. Some Issues in the Classification of DAIS Hyperspectral Data. Int. J. Remote Sens. 2006, 27, 2895–2916. [Google Scholar] [CrossRef]
- Beckschäfer, P. Obtaining Rubber Plantation Age Information from Very Dense Landsat TM & ETM+ Time Series Data and Pixel-Based Image Compositing. Remote Sens. Environ. 2017, 196, 89–100. [Google Scholar] [CrossRef]
- Xie, S.; Liu, L.; Zhang, X.; Yang, J.; Chen, X.; Gao, Y. Automatic Land-Cover Mapping Using Landsat Time-Series Data Based on Google Earth Engine. Remote Sens. 2019, 11, 3023. [Google Scholar] [CrossRef]
- Zeng, L.; Wardlow, B.D.; Xiang, D.; Hu, S.; Li, D. A Review of Vegetation Phenological Metrics Extraction Using Time-Series, Multispectral Satellite Data. Remote Sens. Environ. 2020, 237, 111511. [Google Scholar] [CrossRef]
- Griffiths, P.; van der Linden, S.; Kuemmerle, T.; Hostert, P. A Pixel-Based Landsat Compositing Algorithm for Large Area Land Cover Mapping. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 6, 2088–2101. [Google Scholar] [CrossRef]
- Hermosilla, T.; Wulder, M.A.; White, J.C.; Coops, N.C.; Hobart, G.W. Disturbance-Informed Annual Land cover Classification Maps of Canada’s Forested Ecosystems for a 29-Year Landsat Time Series. Can. J. Remote Sens. 2018, 44, 67–87. [Google Scholar] [CrossRef]
- Archer, K.J.; Kimes, R.V. Empirical Characterization of Random Forest Variable Importance Measures. Comput. Stat. Data Anal. 2008, 52, 2249–2260. [Google Scholar] [CrossRef]
- Rana, V.K.; Venkata Suryanarayana, T.M. Performance Evaluation of MLE, RF and SVM Classification Algorithms for Watershed Scale Land Use/Land Cover Mapping Using Sentinel 2 Bands. Remote Sens. Appl. Soc. Environ. 2020, 19, 100351. [Google Scholar] [CrossRef]
- Mountrakis, G.; Im, J.; Ogole, C. Support Vector Machines in Remote Sensing: A Review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Zhang, Z.; Wei, M.; Pu, D.; He, G.; Wang, G.; Long, T. Assessment of Annual Composite Images Obtained by Google Earth Engine for Urban Areas Mapping Using Random Forest. Remote Sens. 2021, 13, 748. [Google Scholar] [CrossRef]










| Code | Class | Abbreviation | Description |
|---|---|---|---|
| 1 | Others | OT | Other surface types, in addition to the following categories. |
| 2 | Bare land | BL | Areas without vegetation cover, including wasteland, deserts, and the Gobi Desert. |
| 3 | Grassland | GL | Areas where herbaceous plant cover is greater than 15%, including natural grassland and pastures. |
| 4 | Shrubland | SL | Areas in which the shrublands’ height range is 0.3–5 m and cover percentage is >15% have unique texture. |
| 5 | Cropland | CL | It varies greatly throughout the year from bare fields to seeding to crop growing to harvesting. It includes paddy fields, greenhouse agriculture, and other types. |
| 6 | Forest | FO | Areas with tree cover greater than 15% and tree height greater than 3 m. Includes natural forests, planted forests, and fruit trees. |
| Code | Class | Number of Total Samples | Number of Validation Samples |
|---|---|---|---|
| 1 | Others | 7500 | 1500 |
| 2 | Bare land | 10,076 | 2015 |
| 3 | Grassland | 10,550 | 2110 |
| 4 | Shrubland | 10,550 | 2110 |
| 5 | Cropland | 8000 | 1600 |
| 6 | Forest | 7500 | 1500 |
| Dataset | Number of Feature Bands | Description |
|---|---|---|
| 1 | 16 | Median image was composited from April to October of 2020. |
| 2 | 16 | Mean image was composited from April to October of 2020. |
| 3 | 16 | Maximum image was composited from April to October of 2020. |
| 4 | 45 | Median image was composited from April to October and April to July and August to October of 2020. |
| 5 | 52 | Image composited from Dataset 4 and spectral indices of Dataset 3. |
| Land Cover Types | Spectral Bands | Spectral Bands + Spectral Indices | Spectral Bands + Spectral Indices + DEM | ||||||
|---|---|---|---|---|---|---|---|---|---|
| RF | SVM | CART | RF | SVM | CART | RF | SVM | CART | |
| OT | 0.876 | 0.779 | 0.824 | 0.867 | 0.761 | 0.828 | 0.893 | 0.782 | 0.844 |
| BL | 0.933 | 0.856 | 0.829 | 0.935 | 0.879 | 0.836 | 0.96 | 0.876 | 0.872 |
| GL | 0.654 | 0.391 | 0.560 | 0.656 | 0.420 | 0.578 | 0.778 | 0.485 | 0.658 |
| SL | 0.693 | 0.670 | 0.461 | 0.706 | 0.677 | 0.499 | 0.801 | 0.672 | 0.569 |
| CL | 0.670 | 0.490 | 0.480 | 0.708 | 0.513 | 0.510 | 0.770 | 0.526 | 0.593 |
| FO | 0.815 | 0.834 | 0.754 | 0.821 | 0.801 | 0.776 | 0.857 | 0.853 | 0.806 |
| Land Cover Types | Spectral Bands | Spectral Bands + Spectral Indices | Spectral Bands + Spectral Indices + DEM | ||||||
|---|---|---|---|---|---|---|---|---|---|
| RF | SVM | CART | RF | SVM | CART | RF | SVM | CART | |
| OT | 0.914 | 0.813 | 0.830 | 0.924 | 0.824 | 0.824 | 0.942 | 0.826 | 0.854 |
| BL | 0.900 | 0.786 | 0.831 | 0.897 | 0.763 | 0.846 | 0.921 | 0.769 | 0.868 |
| GL | 0.625 | 0.506 | 0.557 | 0.640 | 0.535 | 0.581 | 0.755 | 0.587 | 0.653 |
| SL | 0.701 | 0.534 | 0.445 | 0.714 | 0.557 | 0.461 | 0.792 | 0.552 | 0.545 |
| CL | 0.682 | 0.577 | 0.482 | 0.702 | 0.543 | 0.522 | 0.784 | 0.579 | 0.602 |
| FO | 0.833 | 0.758 | 0.767 | 0.834 | 0.789 | 0.792 | 0.880 | 0.821 | 0.820 |
| OT | BL | GL | SL | CL | FO | Kappa | OA | ||
|---|---|---|---|---|---|---|---|---|---|
| Dataset 1 | PA | 0.893 | 0.960 | 0.778 | 0.801 | 0.770 | 0.857 | 0.801 | 0.835 |
| UA | 0.942 | 0.921 | 0.755 | 0.792 | 0.784 | 0.880 | |||
| Dataset 2 | PA | 0.911 | 0.960 | 0.790 | 0.801 | 0.819 | 0.889 | 0.824 | 0.854 |
| UA | 0.935 | 0.935 | 0.769 | 0.821 | 0.818 | 0.895 | |||
| Dataset 3 | PA | 0.888 | 0.934 | 0.725 | 0.740 | 0.779 | 0.855 | 0.773 | 0.811 |
| UA | 0.909 | 0.915 | 0.716 | 0.772 | 0.765 | 0.843 | |||
| Dataset 4 | PA | 0.903 | 0.982 | 0.820 | 0.842 | 0.846 | 0.922 | 0.855 | 0.880 |
| UA | 0.957 | 0.940 | 0.809 | 0.872 | 0.828 | 0.915 | |||
| Dataset 5 | PA | 0.915 | 0.981 | 0.830 | 0.848 | 0.873 | 0.932 | 0.869 | 0.891 |
| UA | 0.965 | 0.947 | 0.816 | 0.882 | 0.859 | 0.918 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, B.; Yang, L.; Luo, X.; Wu, J.; Hu, L. Extracting Shrubland in Deserts from Medium-Resolution Remote-Sensing Data at Large Scale. Remote Sens. 2024, 16, 374. https://doi.org/10.3390/rs16020374
Zhong B, Yang L, Luo X, Wu J, Hu L. Extracting Shrubland in Deserts from Medium-Resolution Remote-Sensing Data at Large Scale. Remote Sensing. 2024; 16(2):374. https://doi.org/10.3390/rs16020374
Chicago/Turabian StyleZhong, Bo, Li Yang, Xiaobo Luo, Junjun Wu, and Longfei Hu. 2024. "Extracting Shrubland in Deserts from Medium-Resolution Remote-Sensing Data at Large Scale" Remote Sensing 16, no. 2: 374. https://doi.org/10.3390/rs16020374