A Novel Principal Component Analysis Method for the Reconstruction of Leaf Reflectance Spectra and Retrieval of Leaf Biochemical Contents
Abstract
:1. Introduction
2. Materials and Methods
2.1. Measured and Simulated Datasets
2.1.1. LOPEX93 Dataset
2.1.2. ANGERS Dataset
2.1.3. Simulated PROSPECT Model Dataset
2.2. Vegetation Indices Sensitive to Leaf Biochemical Contents
2.3. Principal Component Analysis
3. Results
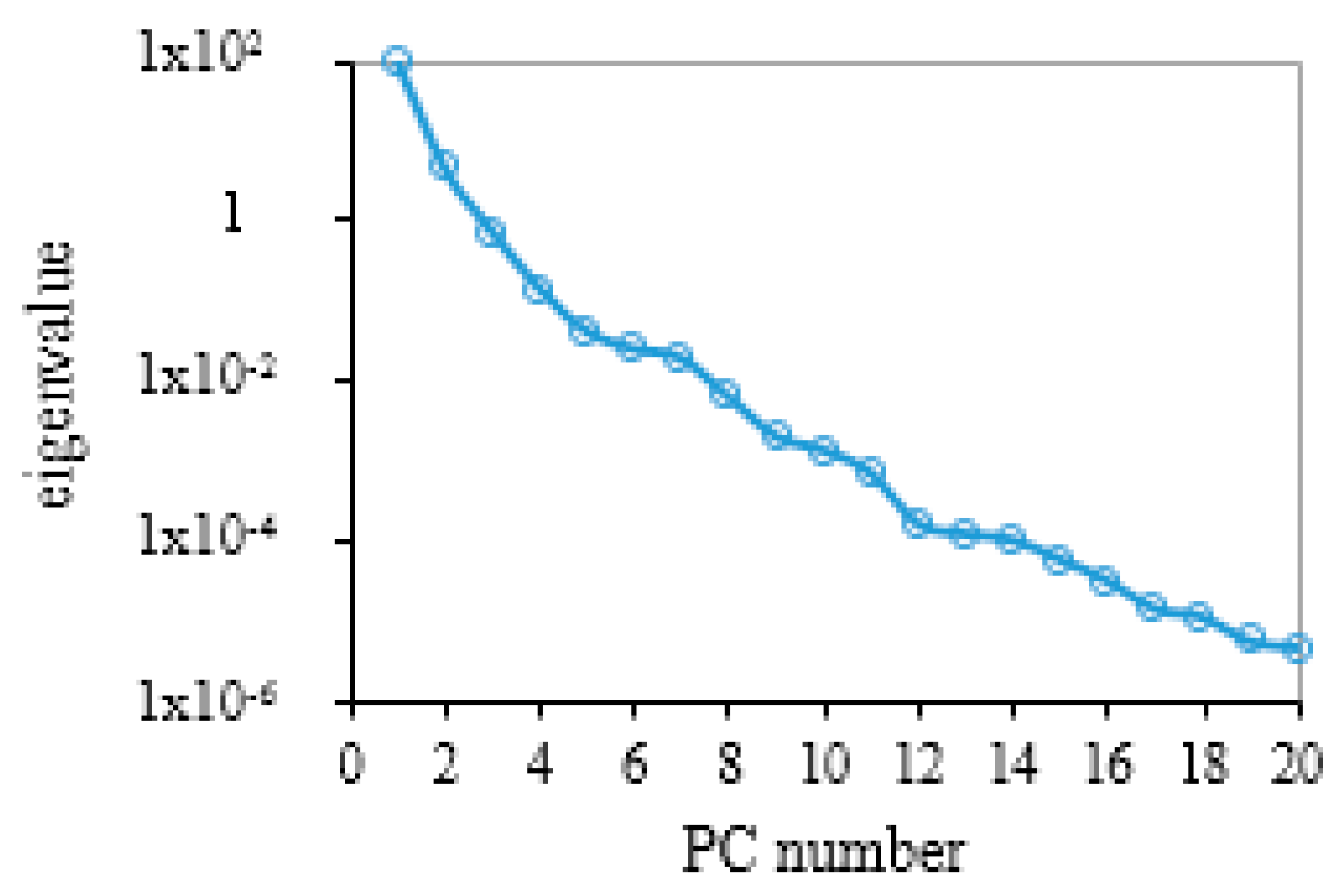
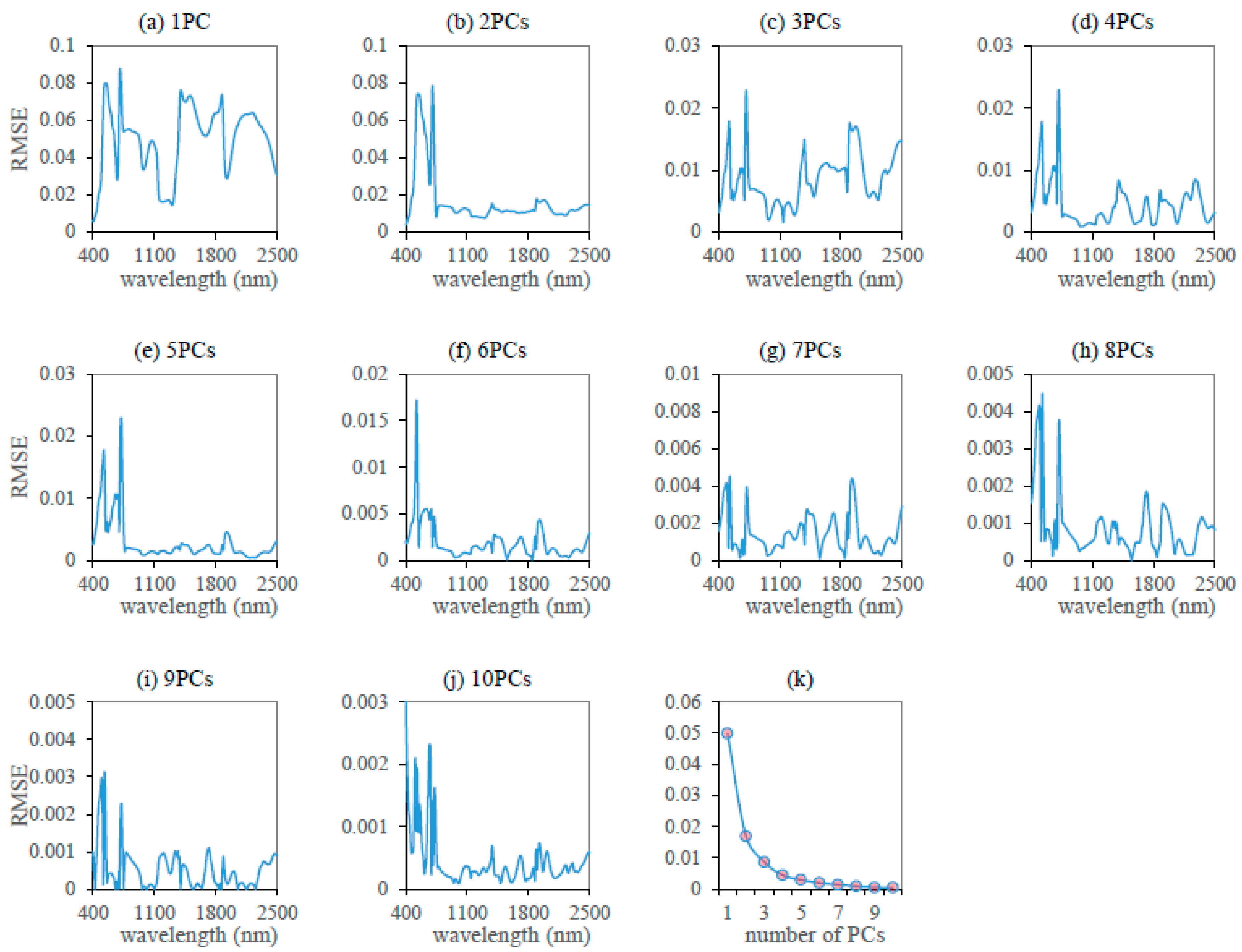
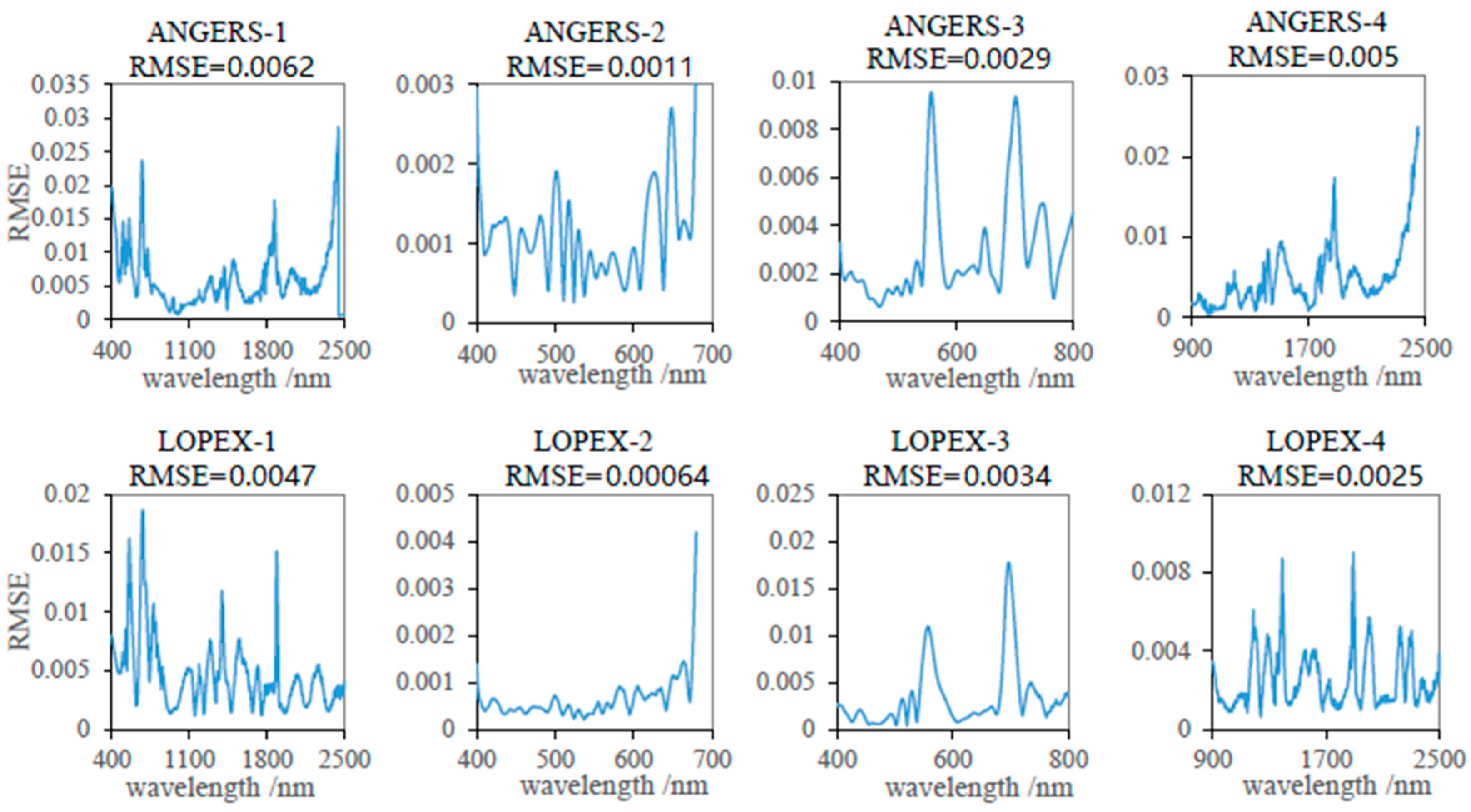
3.1. Reconstruction of the Leaf Reflectance Spectra Using the PCA Data-Driven Method
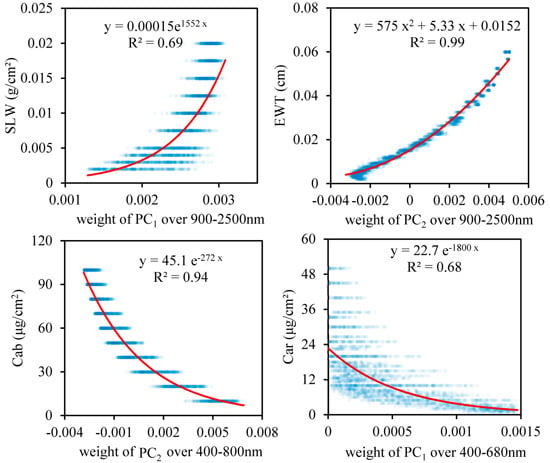
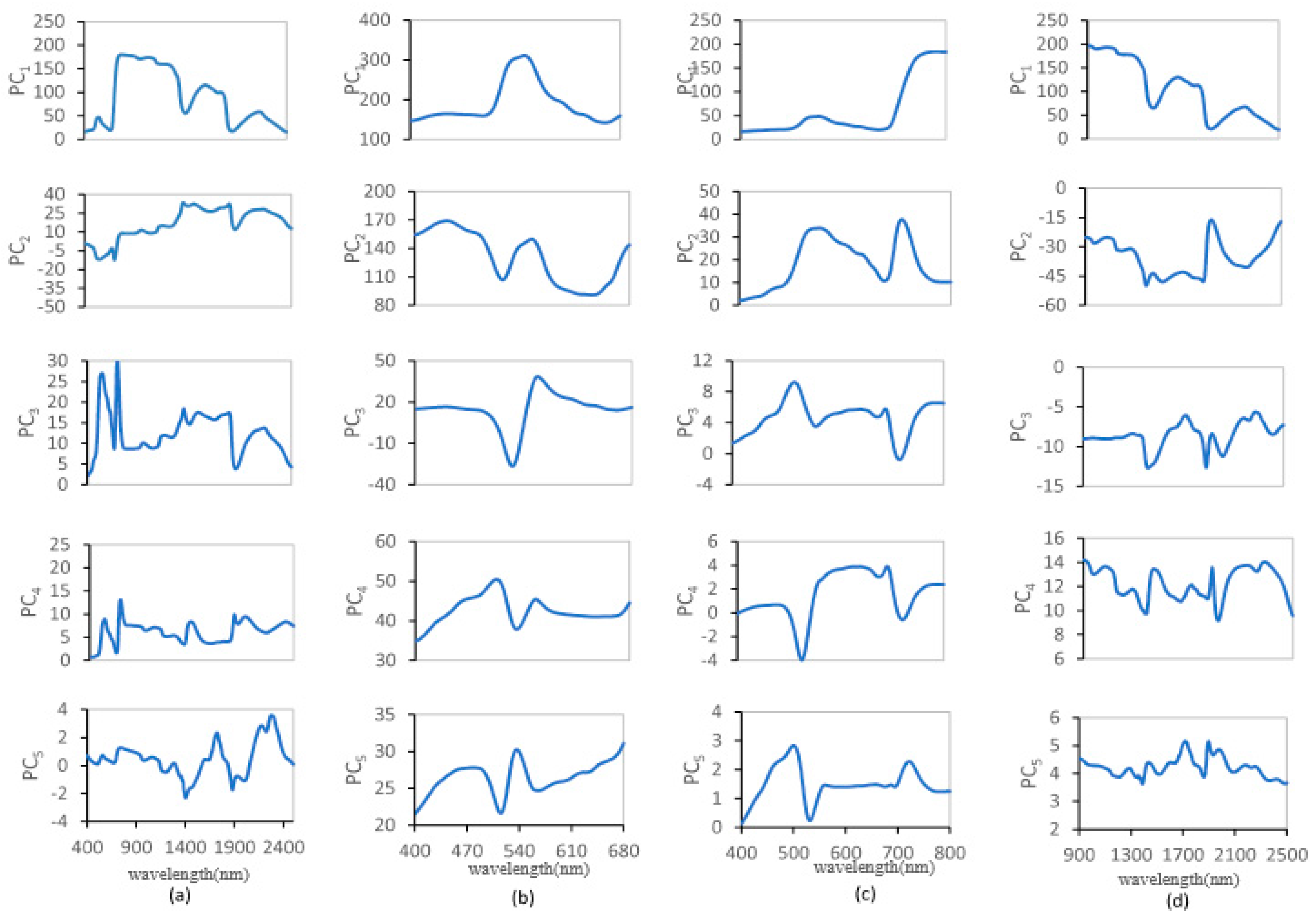
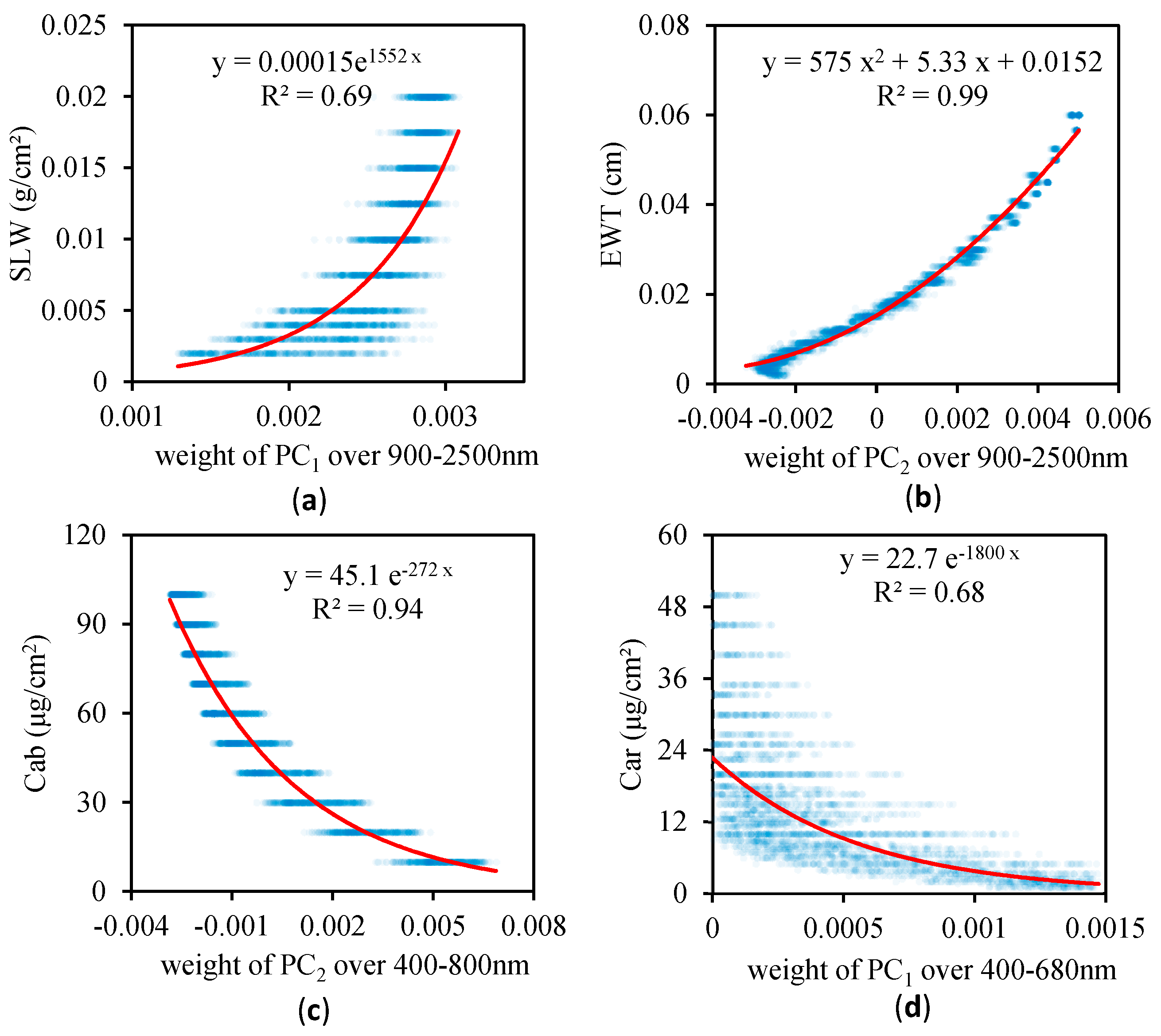
3.2. Relationship between the Weighting Coefficients of the PCs and the Leaf Biochemical Contents
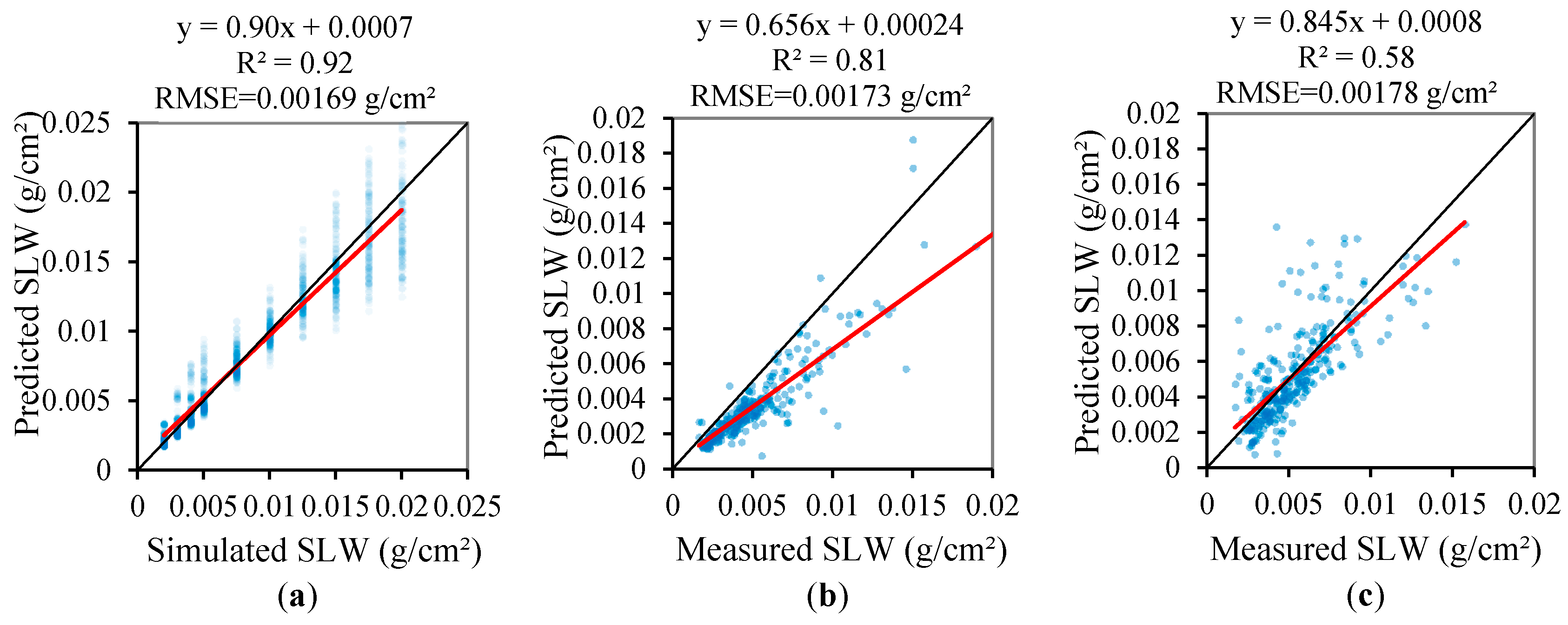
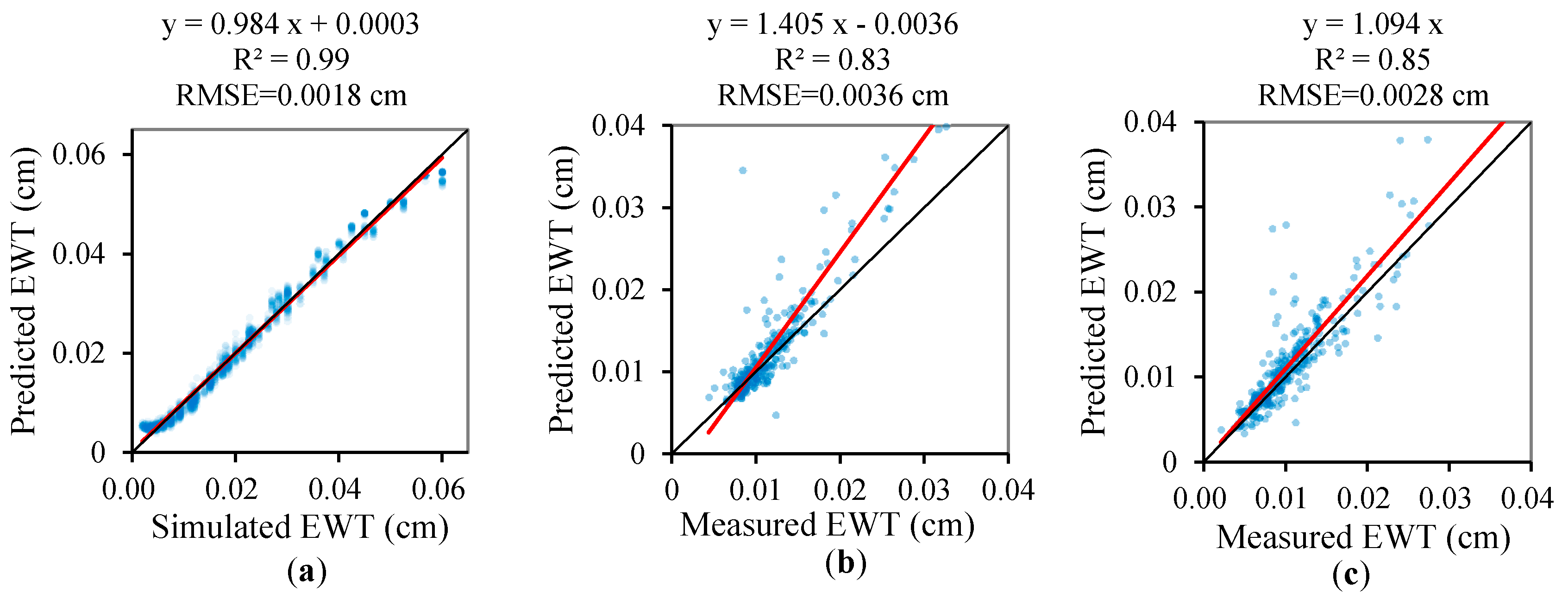
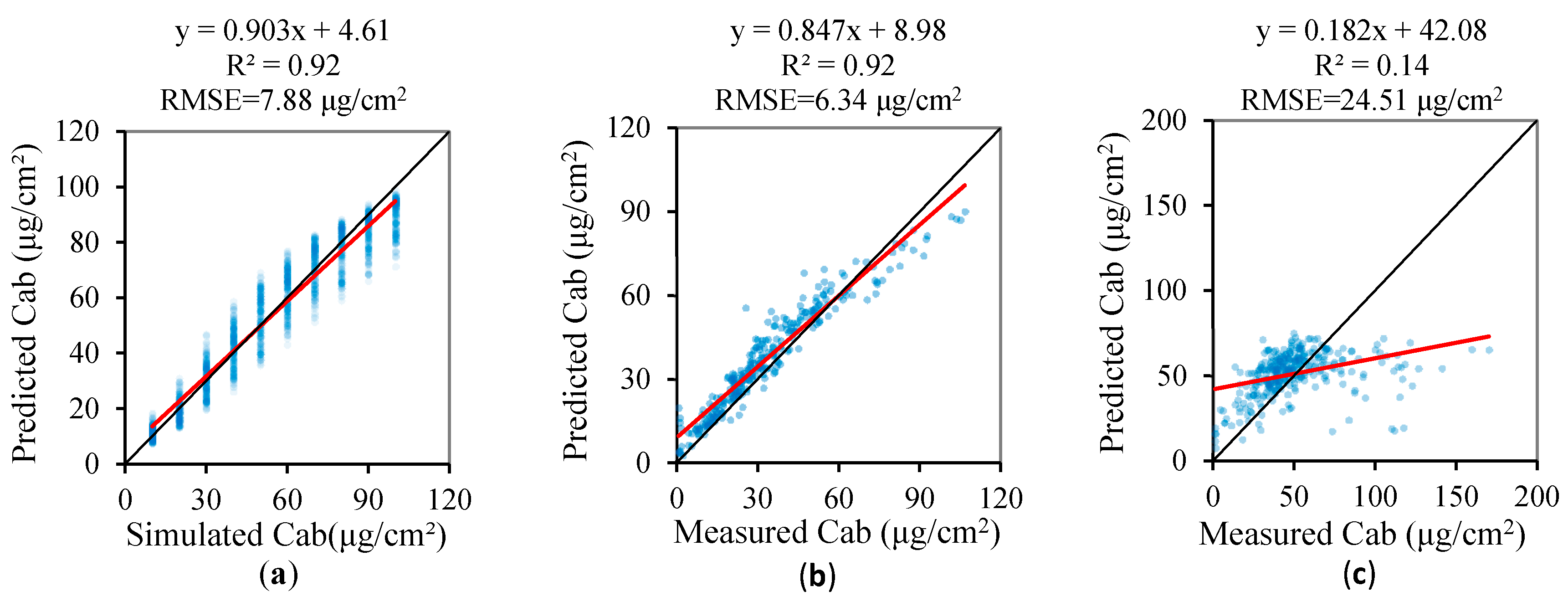
3.3. Validation of the Regression Models of the Leaf Biochemical Contents
3.4. Comparison of PCA Method with Traditional VI-Based Method for Retrieving Leaf Biochemical Contents
- (1)
- the SLW can be accurately retrieved using the NDMI and NDLMA, and the PCA method also only showed similar or slightly better performance on estimation for ANGERS and LOPEX’93.
- (2)
- the EWT can be more accurately retrieved using the PCA method than those obtained using the VI-based method for all validation datasets.
- (3)
- the PCA method generally provides a better estimate of the pigment content, although the ND705–based and CIred edge–based models provide a slightly more accurate estimation for Cab when the ANGERS is used.
4. Discussion
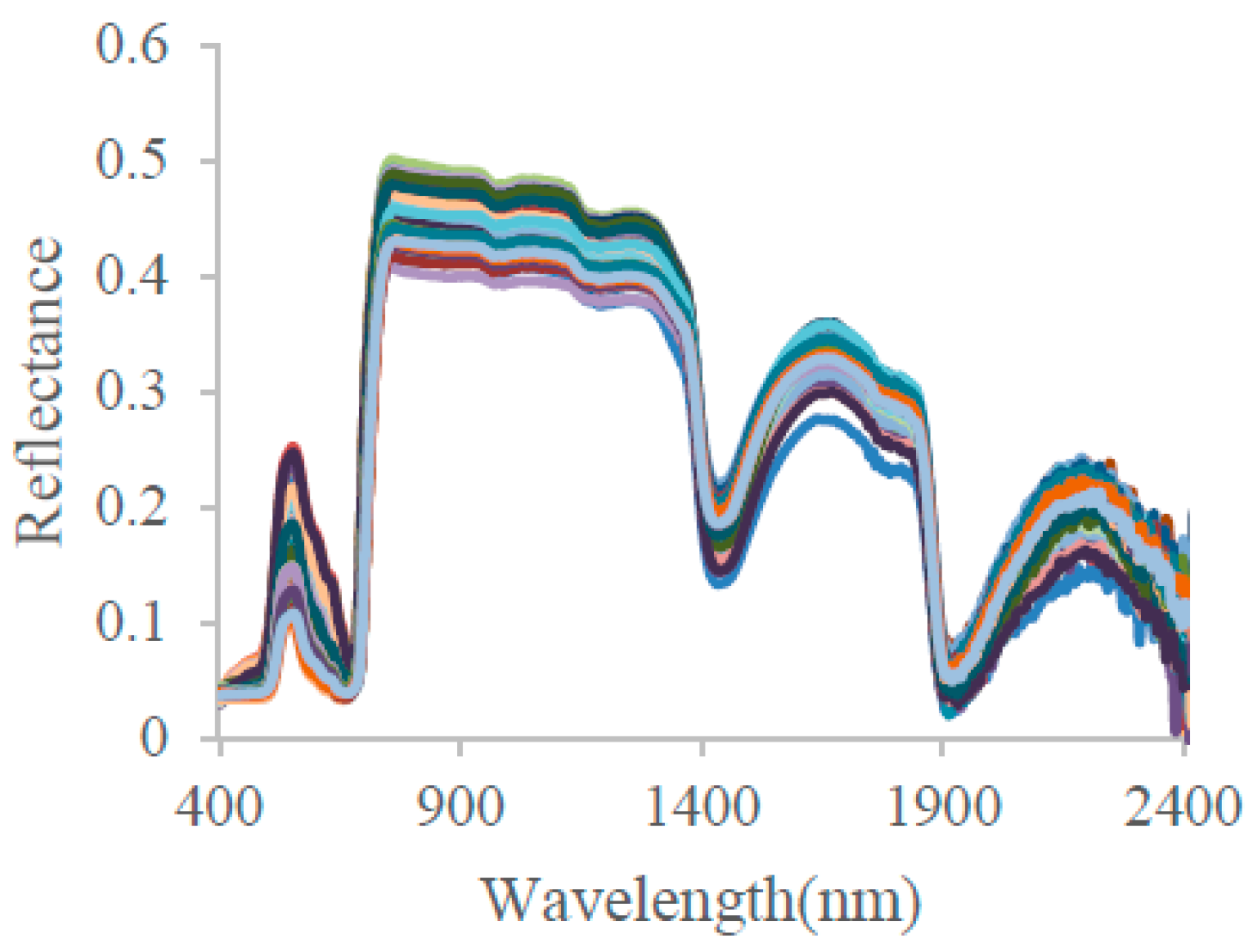
4.1. The Performance of the PCA Approach in the Reconstruction of Leaf Reflectance Spectra
4.2. The Performance of the PCA Approach in the Estimation of Leaf Parameters
4.3. Limitations of the PCA Data-Driven Method
5. Conclusions
- ◆
- The leaf reflectance spectra can be accurately linearly reconstructed using only a few leading PCs, with the ten leading PCs accounting for 99.998% of the total information contained in the 3636 training data. The spectral RMSE between measured reflectance and reconstructed reflectance using the PCA method was found to be about 3–10 times smaller than that using the PROSPECT simulation method for the measured datasets. Therefore, the PCA method may provide a new data-driven approach for the reconstruction of leaf reflectance spectra.
- ◆
- The spectra of some of the leading PCs are similar to the contributions of the leaf biochemical components to the reflectance, and the weighting coefficients of the PCs are significantly correlated with the leaf biochemical contents. If only the weighting coefficient of the most sensitive PC was employed, the coefficients of determination for the PCA data-driven model were 0. 69, 0.99, 0.94 and 0.68 for SLW, EWT, Cab and Car, respectively.
- ◆
- The PCA data-driven models were validated and compared to the traditional VI-based and physical approaches to the retrieval of leaf properties. The results show that the PCA method gives similar or even better estimation of most of the leaf biochemical contents, including the SLW, EWT, Cab and Car. Therefore, the PCA data-driven method also provides a new way of retrieving leaf biochemical contents that may be more robust and accurate than the traditional VI-based and full-physical methods.
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Clevers, J.G.P.W. The use of imaging spectrometry for agricultural applications. ISPRS J. Photogramm. Remote Sens. 1999, 54, 299–304. [Google Scholar] [CrossRef]
- Cox, S. Information technology: The global key to precision agriculture and sustainability. Comput. Electron. Agric. 2002, 36, 93–111. [Google Scholar] [CrossRef]
- Potter, C.S.; Klooster, S.; Brooks, V. Interannual variability in terrestrial net primary production: Exploration of trends and controls on regional to global scales. Ecosystems 1999, 2, 36–48. [Google Scholar] [CrossRef]
- Ustin, S.L. Remote sensing of canopy chemistry. Proc. Natl. Acad. Sci. USA 2003, 110, 804–805. [Google Scholar] [CrossRef] [PubMed]
- Bacour, C.; Baret, F.; Béal, D.; Weiss, M.; Pavageau, K. Neural network estimation of LAI, fAPAR, fCover, and LAI × Cab, from top of canopy MERIS reflectance data: Principles and validation. Remote Sens. Environ. 2006, 105, 313–325. [Google Scholar] [CrossRef]
- Dorigo, W.A.; Zurita-Milla, R.; de Wit, A.J.W.; Brazile, J.; Singh, R.; Schaepman, M.E. A review on reflective remote sensing and data assimilation techniques for enhanced agroecosystem modeling. Int. J. Appl. Earth Obs. Geoinf. 2007, 9, 165–193. [Google Scholar] [CrossRef]
- Verrelst, J.; Camps-Valls, G.; Muñoz-Marí, J.; Rivera, J.P.; Veroustraete, F.; Clevers, J.G.; Moreno, J. Optical remote sensing and the retrieval of terrestrial vegetation bio-geophysical properties—A review. ISPRS J. Photogramm. Remote Sens. 2015, 108, 273–290. [Google Scholar] [CrossRef]
- Dorigo, W.A.; Richter, R.; Baret, F.; Bamler, R.; Wagner, W. Enhanced automated canopy characterization from hyperspectral data by a novel two step radiative transfer model inversion approach. Remote Sens. 2009, 1, 1139–1170. [Google Scholar] [CrossRef]
- Downing, H.G.; Carter, G.A.; Holladay, K.W.; Cibula, W.G. The radiative-equivalent water thickness of leaves. Remote Sens. Environ. 1993, 46, 103–107. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Hunt, E.R.; Rock, B.N. Detection of changes in leaf water content using near and middle-infrared reflectances. Remote Sens. Environ. 1989, 30, 43–54. [Google Scholar]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Harris, A.; Dash, J. The potential of the MERIS Terrestrial Chlorophyll index for carbon flux estimation. Remote Sens. Environ. 2010, 114, 1856–1862. [Google Scholar] [CrossRef]
- Liu, L.Y.; Huang, W.J.; PU, R.L.; Wang, J.H. Detection of internal leaf structure deterioration using a new spectral ratio index in the near-infrared shoulder region. J. Integr. Agric. 2014, 13, 760–769. [Google Scholar] [CrossRef]
- Miller, J.R.; Hare, E.W.; Wu, J. Quantitative characterization of the vegetation red edge reflectance 1. An inverted-Gaussian reflectance model. Remote Sens. 1990, 11, 1755–1773. [Google Scholar] [CrossRef]
- Filella, I.; Peñuelas, J. The red edge position and shape as indicators of plant chlorophyll content, biomass and hydric status. Int. J. Remote Sens. 1994, 15, 1459–1470. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N.; Lichtenthaler, H.K. Detection of Red Edge Position and Chlorophyll Content by Reflectance Measurements near 700 nm. J. Plant Physiol. 1996, 148, 501–508. [Google Scholar] [CrossRef]
- Liu, L.Y.; Wang, J.H.; Huang, W.J.; Zhao, C.J.; Zhang, B.; Tong, Q.X. Estimating winter wheat plant water content using red edge parameters. Int. J. Remote Sens. 2004, 25, 3331–3342. [Google Scholar] [CrossRef]
- Kokaly, R.F.; Clark, R.N. Spectroscopic determination of leaf biochemistry using band-depth analysis of absorption features and stepwise multiple linear regression. Remote Sens. Environ. 1999, 67, 267–287. [Google Scholar] [CrossRef]
- Pu, R.L.; Gong, P.; Biging, G.S.; Larrieu, M.R. Extraction of Red Edge Optical Parameters from Hyperion Data for Estimation of Forest Leaf Area Index. IEEE Trans. Geosci. Remote Sens. 2003, 41, 916–921. [Google Scholar]
- Zarco-Tejada, P.J.; Miller, J.R.; Mohammed, G.H.; Noland, T.L.; Sampson, P.H. Vegetation stress detection through chlorophyll + estimation and fluorescence effects on hyperspectral imagery. J. Environ. Qual. 2002, 31, 1433–1441. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.Y.; Wang, J.H.; Huang, W.J.; Zhao, C.J. Detection of leaf and canopy EWT by calculating REWT from reflectance spectra. Int. J. Remote Sens. 2010, 31, 2681–2695. [Google Scholar] [CrossRef]
- Pu, R.L.; Gong, P. Wavelet tansform applied to EO-1 hyperspectral data for forest LAI and crown closure mapping. Remote Sens. Environ. 2004, 91, 212–224. [Google Scholar] [CrossRef]
- Atzberger, C.; Guérif, M.; Baret, F.; Werner, W. Comparative analysis of three chemometric techniques for the spectroradiometric assessment of canopy chlorophyll content in winter wheat. Comput. Electron. Agric. 2010, 73, 165–173. [Google Scholar] [CrossRef]
- Cheng, T.; Rivard, B.; Sanchez-Azofeifa, A. Spectroscopic determination of leaf water content using continuous wavelet analysis. Remote Sens. Environ. 2011, 115, 659–670. [Google Scholar] [CrossRef]
- Cheng, T.; Rivard, B.; Sánchez-Azofeifa, G.A.; Féret, J.B.; Jacquemoud, S.; Ustin, S.L. Deriving leaf mass per area (LMA) from foliar reflectance across a variety of plant species using continuous wavelet analysis. ISPRS J. Photogramm. Remote Sens. 2014, 87, 28–38. [Google Scholar] [CrossRef]
- Knyazikhin, Y.; Schull, M.A.; Stenberg, P.; Mõttus, M.; Rautiainen, M.; Yang, Y.; Marshak, A.; Carmona, L.P.; Kaufmann, R.K.; Lewis, P.; et al. Hyperspectral remote sensing of foliar nitrogen content. Proc. Natl. Acad. Sci. USA 2013, 110, E185–E192. [Google Scholar] [CrossRef] [PubMed]
- Jacquemoud, S.; Verhoef, W.; Baret, F.; Bacour, C.; Zarco-Tejada, P.J.; Asner, G.P.; Franҫois, C.; Ustin, S.L. PROSPECT + SAIL models: A review of use for vegetation characterization. Remote Sens. Environ. 2009, 113, S56–S66. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Baret, F.; Andrieu, B.; Danson, F.M.; Jaggard, K. Extraction of vegetation biophysical parameters by inversion of the PROSPECT+SAIL models on sugar beet canopy reflectance data. Application to TM and AVIRIS sensors. Remote Sens. Environ. 1995, 52, 163–172. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Ustin, S.L.; Verdebout, J.; Schmuck, G.; Andreoli, G.; Hosgood, B. Estimating leaf biochemistry using the PROSPECT leaf optical properties model. Remote Sens. Environ. 1996, 56, 194–202. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Bacour, C.; Poilvé, H.; Frangi, J.P. Comparison of four radiative transfer models to simulate plant canopies reflectance: Direct and inverse mode. Remote Sens. Environ. 2000, 74, 471–481. [Google Scholar] [CrossRef]
- Combal, B.; Baret, F.; Weiss, M.; Trubuil, A.; Macé, D.; Pragnère, A.; Myneni, P.; Knyazikhin, Y.; Wang, L. Retrieval of canopy biophysical variables from bidirectional reflectance: Using prior information to solve the ill-posed inverse problem. Remote Sens. Environ. 2002, 84, 1–15. [Google Scholar] [CrossRef]
- Weiss, M.; Baret, F.; Myneni, R.B.; Pragnère, A.; Knyazikhin, Y. Investigation of a model inversion technique to estimate canopy biophysical variables from spectral and directional reflectance data. Agronomie 2000, 20, 3–22. [Google Scholar] [CrossRef]
- Riaño, D.; Vaughan, P.; Chuvieco, E.; Zarco-Tejada, P.J.; Ustin, S.L. Estimation of fuel moisture content by inversion of radiative transfer models to simulate equivalent water thickness and dry matter content: Analysis at leaf and canopy level. IEEE Trans. Geosci. Remote Sens. 2005, 43, 819–826. [Google Scholar] [CrossRef]
- Weiss, M.; Baret, F. Evaluation of canopy biophysical variable retrieval performances from the accumulation of large swath satellite data. Remote Sens. Environ. 1999, 70, 293–306. [Google Scholar] [CrossRef]
- Walthall, C.; Dulaney, W.; Anderson, M.; Norman, J.; Fang, H.; Liang, S. A comparison of empirical and neural network approaches for estimating corn and soybean leaf area index from Landsat ETM + imagery. Remote Sens. Environ. 2004, 92, 465–474. [Google Scholar] [CrossRef]
- Fang, H.; Liang, S. A hybrid inversion method for mapping leaf area index from MODIS data: Experiments and application to broadleaf and needleleaf canopies. Remote Sens. Environ. 2005, 94, 405–424. [Google Scholar] [CrossRef]
- Baret, F.; Hagolle, O.; Geiger, B.; Bicheron, P.; Miras, B.; Huc, M.; Berthelot, B.; Niño, F.; Weiss, M.; Samain, O.; et al. LAI, fAPAR and fCover CYCLOPES global products derived from VEGETATION: Part 1: Principles of the algorithm. Remote Sens. Environ. 2007, 110, 275–286. [Google Scholar] [CrossRef] [Green Version]
- Verrelst, J.; Munoz, J.; Alonso, L.; Delegido, J.; Pablo Rivera, J.; Camps-Valls, G.; Moreno, J. Machine learning regression algorithms for biophysical parameter retrieval: Opportunities for Sentinel-2 and -3. Remote Sens. Environ. 2012, 118, 127–139. [Google Scholar] [CrossRef]
- Campos-Taberner, M.; García-Haro, F.J.; Camps-Valls, G.; Grau-Muedra, G.; Nutini, F.; Crema, A.; Boschetti, M. Multitemporal and multiresolution leaf area index retrieval for operational local rice crop monitoring. Remote Sens. Environ. 2016, 187, 102–118. [Google Scholar] [CrossRef]
- Campos-Taberner, M.; García-Haro, F.J.; Moreno, A.; Gilabert, M.A.; Sanchez-Ruiz, S.; Martinez, B.; Camps-Valls, G. Mapping leaf area index with a smartphone and Gaussian processes. IEEE Geosci. Remote Sens. Lett. 2015, 12, 2501–2505. [Google Scholar] [CrossRef]
- Lázaro-Gredilla, M.; Titsias, M.K.; Verrelst, J.; Camps-Valls, G. Retrieval of biophysical parameters with heteroscedastic Gaussian processes. IEEE Geosci. Remote Sens. Lett. 2014, 11, 838–842. [Google Scholar] [CrossRef]
- Rivera-Caicedo, J.P.; Verrelst, J.; Muñoz-Marí, J.; Camps-Valls, G.; Moreno, J. Hyperspectral dimensionality reduction for biophysical variable statistical retrieval. ISPRS J. Photogramm. Remote Sens. 2017, 132, 88–101. [Google Scholar] [CrossRef]
- Bréda, N.J. Ground-based measurements of leaf area index: A review of methods, instruments and current controversies. J. Exp. Bot. 2003, 54, 2403–2417. [Google Scholar] [CrossRef] [PubMed]
- Jonckheere, I.; Fleck, S.; Nackaerts, K.; Muys, B.; Coppin, P.; Weiss, M.; Baret, F. Review of methods for in situ leaf area index determination: Part I. Theories, sensors and hemispherical photography. Agric. For. Meteorol. 2004, 121, 19–35. [Google Scholar] [CrossRef]
- Weiss, M.; Baret, F.; Smith, G.J.; Jonckheere, I.; Coppin, P. Review of methods for in situ leaf area index (LAI) determination: Part II. Estimation of LAI, errors and sampling. Agric. For. Meteorol. 2004, 121, 37–53. [Google Scholar] [CrossRef]
- Chen, G.; Qian, S.E. Denoising of hyperspectral imagery using principal component analysis and wavelet shrinkage. IEEE Trans. Geosci. Remote Sens. 2011, 49, 973–980. [Google Scholar] [CrossRef]
- Keshava, N.; Mustard, J.F. Spectral unmixing. IEEE Signal Process. Mag. 2002, 19, 44–57. [Google Scholar] [CrossRef]
- Farrell, M.D.; Mersereau, R.M. On the impact of PCA dimension reduction for hyperspectral detection of difficult targets. IEEE Geosci. Remote Sens. Lett. 2005, 2, 192–195. [Google Scholar] [CrossRef]
- Chaurasia, S.; Dadhwal, V.K. Comparison of principal component inversion with VI-empirical approach for LAI estimation using simulated reflectance data. Int. J. Remote Sens. 2004, 25, 2881–2887. [Google Scholar] [CrossRef]
- Satapathy, S.; Dadhwal, V.K. Principal component inversion technique for the retrieval of leaf area index. J. Indian Soc. Remote Sens. 2005, 33, 323–330. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Ma, X.M.; Jia, F.F.; Qiao, H.B.; Zhang, Y.W. Hyperspectral estimating models of tobacco leaf area index. Acta Ecol. Sin. 2012, 32, 168–175. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Skidmore, A.; Schlerf, M.; Atzberger, C.; Corsi, F.; Cho, M. LAI and chlorophyll estimation for a heterogeneous grassland using hyperspectral measurements. ISPRS J. Photogramm. Remote Sens. 2008, 63, 409–426. [Google Scholar] [CrossRef]
- Gonsamo, A.; Pellikka, P.; King, D.J. Large-scale leaf area index inversion algorithms from high-resolution airborne imagery. Int. J. Remote Sens. 2011, 32, 3897–3916. [Google Scholar] [CrossRef]
- Ray, S.S.; Das, G.; Singh, J.P.; Panigrahy, S. Evaluation of hyperspectral indices for LAI estimation and discrimination of potato crop under different irrigation treatments. Int. J. Remote Sens. 2006, 27, 5373–5387. [Google Scholar] [CrossRef]
- Li, Z.W.; Xin, X.P.; Huan, T.A.N.G.; Fan, Y.A.N.G.; Chen, B.R.; Zhang, B.H. Estimating grassland LAI using the Random Forests approach and Landsat imagery in the meadow steppe of Hulunber, China. J. Integr. Agric. 2017, 16, 286–297. [Google Scholar] [CrossRef]
- Feng, R.; Zhang, Y.; Yu, W.; Hu, W.; Wu, J.; Ji, R.; Zhao, X. Analysis of the relationship between the spectral characteristics of maize canopy and leaf area index under drought stress. Acta Ecol. Sin. 2013, 33, 301–307. [Google Scholar] [CrossRef]
- Atzberger, C.; Jarmer, T.; Schlerf, M.; Kötz, B.; Werner, W. Spectroradiometric determination of wheat bio-physical variables: Comparison of different empirical-statistical approaches. In Proceedings of the 23rd EARSeL Symposium and General Assembly: Remote Sensing in Transition, Ghent, Belgium, 2–5 June 2003; pp. 2–5. [Google Scholar]
- Zhang, J.; Han, W.; Huang, L.; Zhang, Z.; Ma, Y.; Hu, Y. Leaf chlorophyll content estimation of winter wheat based on visible and near-infrared sensors. Sensors 2016, 16, 437. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.W.; Laird, D.A.; Mausbach, M.J.; Hurburgh, C.R. Near-infrared reflectance spectroscopy–principal components regression analyses of soil properties. Soil Sci. Soc. Am. J. 2001, 65, 480–490. [Google Scholar] [CrossRef]
- Çamdevýren, H.; Demýr, N.; Kanik, A.; Keskýn, S. Use of principal component scores in multiple linear regression models for prediction of Chlorophyll-a in reservoirs. Ecol. Model. 2005, 181, 581–589. [Google Scholar] [CrossRef]
- Zhou, L.; Ma, W.; Zhang, H.; Li, L.; Tang, L. Developing a PCA–ANN Model for Predicting Chlorophyll a Concentration from Field Hyperspectral Measurements in Dianshan Lake, China. Exposure Health 2015, 7, 591–602. [Google Scholar] [CrossRef]
- Rabbette, M.; Pilewskie, P. Multivariate analysis of solar spectral irradiance measurements. J. Geophys. Res. Atmos. 2001, 106, 9685–9696. [Google Scholar] [CrossRef]
- Huang, H.L.; Antonelli, P. Application of principal component analysis to high-resolution infrared measurement compression and retrieval. J. Appl. Meteorol. 2001, 40, 365–388. [Google Scholar] [CrossRef]
- Joiner, J.; Guanter, L.; Lindstrot, R.; Voigt, M.; Vasilkov, A.P.; Middleton, E.M.; Huemmrich, K.F.; Yoshida, Y.; Frankenberg, C. Global monitoring of terrestrial chlorophyll fluorescence from moderate-spectral-resolution near-infrared satellite measurements: Methodology, simulations, and application to GOME-2. Atmos. Meas. Tech. 2013, 6, 2803–2823. [Google Scholar] [CrossRef]
- Guanter, L.; Aben, I.; Tol, P.; Krijger, J.M.; Hollstein, A.; Köhler, P.; Damm, A.; Joiner, J.; Frankenberg, C.; Landgraf, J. Potential of the TROPOspheric monitoring instrument (TROPOMI) onboard the sentinel-5 precursor for the monitoring of terrestrial chlorophyll fluorescence. Atmos. Meas. Tech. 2014, 8, 1337–1352. [Google Scholar] [CrossRef] [Green Version]
- Köhler, P.; Guanter, L.; Joiner, J. A linear method for the retrieval of sun-induced chlorophyll fluorescence from GOME-2 and SCIAMACHY data. Atmos. Meas. Tech. 2015, 8, 2589–2608. [Google Scholar] [CrossRef]
- Li, C.; Joiner, J.; Krotkov, N.A.; Bhartia, P.K. A fast and sensitive new satellite SO2 retrieval algorithm based on principal component analysis: Application to the ozone monitoring instrument. Geophys. Res. Lett. 2013, 40, 6314–6318. [Google Scholar] [CrossRef]
- Hosgood, B.; Jacquemoud, S.; Andreoli, G.; Verdebout, J.; Pedrini, G.; Schmuck, G. Leaf Optical Properties EXperiment 93 (LOPEX93); European Commission—Joint Research Centre: Ispra, Italy, 1994; Available online: https://ec.europa.eu/jrc/en/about/jrc-site/ispra (accessed on 21 September 2017).
- The Database on Leaf Optical Properties. Available online: http://opticleaf.ipgp.fr/index.php?page=database (accessed on 21 September 2017).
- Féret, J.B.; François, C.; Asner, G.P.; Gitelson, A.A.; Martin, R.; Bidel, L.P.R.; Ustin, S.L.; Maire, G.L.; Jacquemoud, S. PROSPECT-4 and 5: Advances in the leaf optical properties model separating photosynthetic pigments. Remote Sens. Environ. 2008, 112, 3030–3043. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Baret, F. PROSPECT: A model of leaf optical properties spectra. Remote Sens. Environ. 1990, 34, 75–91. [Google Scholar] [CrossRef]
- Bacour, C.; Jacquemoud, S.; Tourbier, Y.; Dechambre, M.; Frangi, J.P. Design and analysis of numerical experiments to compare four canopy reflectance models. Remote Sens. Environ. 2002, 79, 72–83. [Google Scholar] [CrossRef]
- Aldakheel, Y.Y.; Danson, F.M. Spectral reflectance of dehydrating leaves: Measurements and modelling. Int. J. Remote Sens. 1997, 18, 3683–3690. [Google Scholar] [CrossRef]
- Dawson, T.P.; Curran, P.J.; North, P.R.J.; Plummer, S.E. The propagation of foliar biochemical absorption features in forest canopy reflectance: A theoretical analysis. Remote Sens. Environ. 1999, 67, 147–159. [Google Scholar] [CrossRef]
- Rouse, J.W. Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation; NASA/GSFC, Type III, Final Report; NASA: Greenbelt, MD, USA, 1974; pp. 1–371.
- Pearson, R.L.; Miller, L.D. Remote mapping of standing crop biomass for estimation of the productivity of the shortgrass prairie. In Proceedings of the Eighth International Symposium on Remote Sensing of Environment, Ann Arbor, MI, USA, 2–6 October 1972; p. 1355. [Google Scholar]
- Dash, J.; Curran, P.J. The MERIS terrestrial chlorophyll index. Int. J. Remote Sens. 2004, 25, 5003–5013. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Gamon, J.A.; Penuelas, J.; Field, C.B. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Viña, A.; Rundquist, D.C.; Ciganda, V.; Arkebauer, T.J. Remote estimation of canopy chlorophyll content in crops. Geophys. Res. Lett. 2005, 32, L08403. [Google Scholar] [CrossRef]
- Peñuelas, J.; Baret, F.; Filella, I. Semi-empirical indices to assess carotenoids/chlorophyll a ratio from leaf spectral reflectance. Photosynthetica 1995, 31, 221–230. [Google Scholar]
- Gao, B.C. NDWI—A normalized difference water index for remote sensing of vegetation liquid water from space. Remote Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Peñuelas, J.; Pinol, J.; Ogaya, R.; Filella, I. Estimation of plant water concentration by the reflectance Water Index WI (R900/R970). Int. J. Remote Sens. 1997, 18, 2869–2875. [Google Scholar] [CrossRef]
- Féret, J.B.; François, C.; Gitelson, A.; Asner, G.P.; Barry, K.M.; Panigada, C.; Richardson, A.D.; Jacquemoud, S. Optimizing spectral indices and chemometric analysis of leaf chemical properties using radiative transfer modeling. Remote Sens. Environ. 2011, 115, 2742–2750. [Google Scholar] [CrossRef]
- Wang, L.; Hunt, E.R.; Qu, J.J.; Hao, X.; Daughtry, C.S. Towards estimation of canopy foliar biomass with spectral reflectance measurements. Remote Sens. Environ. 2011, 115, 836–840. [Google Scholar] [CrossRef]
- Ceccato, P.; Flasse, S.; Tarantola, S.; Jacquemoud, S.; Grégoire, J.M. Detecting vegetation water content using reflectance in the optical domain. Remote Sens. Environ. 2001, 77, 22–33. [Google Scholar]
- Jacquemoud, S.; Ustin, L.S. Modeling Leaf Optical Properties. Photobiological Sciences Online. 2008. Available online: http://photobiology.info/Jacq_Ustin.html (accessed on 21 September 2017).
- Féret, J.B.; Gitelson, A.A.; Noble, S.D.; Jacquemoud, S. PROSPECT-D: Towards modeling leaf optical properties through a complete lifecycle. Remote Sens. Environ. 2017, 193, 204–215. [Google Scholar] [CrossRef]

| Variable | Values | Unit | Description |
|---|---|---|---|
| Cab | 10–100 with an interval of 10 | μg/cm2 | Chlorophyll content |
| SLW | [0.002, 0.003, 0.004, 0.005, 0.0075, 0.01, 0.0125, 0.015, 0.0175, 0.02] | g/cm2 | Specific leaf weight |
| EWT | Derived from gravimetric water content (GWC %) and SLW GWC: 50–90% with an interval of 5% | cm | Equivalent water thickness |
| Car | [1/10, 1/8, 1/6, 1/5, 1/4, 1/3, 1/2] × Cab | μg/cm2 | Carotenoid content |
| N | Determined according to SLW | - | Leaf structure parameter |
| Biochemical Component | Spectral Index | Formula | Reference |
|---|---|---|---|
| chlorophyll/Carotenoid | NDVI | NDVI = (R800 − R670)/(R800 + R670) | Rouse [76] |
| RVI | RVI = R800/R670 | Pearson and Miller [77] | |
| MTCI | MTCI = (R750 − R710)/(R710 − R680) | Dash and Curran [78] | |
| ND705 | (R750 − R705)/(R750 + R705) | Sims and Gamon [79] | |
| PRI | (R531 − R570)/(R531 + R570) | Gamon et al. [80] | |
| CIgreen | (RNIR/Rgreen) − 1 | Gitelson et al. [10] | |
| CIred edge | (RNIR/Rred edge) − 1 | Gitelson et al. [81] | |
| SIPI | (R800 − R445)/(R800 − R680) | Peñuelas et al. [82] | |
| Water | NDWI | NDWI = (R860 − R1240)/(R860 + R1240) | Gao [83] |
| WI | WI = R900/R970 | Peñuelas et al. [84] | |
| MSI | MSI = R1600/R820 | Hunt et al. [11] | |
| SLW | NDLMA | (R1368 − R1722)/(R1368 + R1722) | Féret et al. [85] |
| NDMI | (R1649 − R1722)/(R1649 + R1722) | Wang et al. [86] |
| Spectral Region (nm) | Training | Validation | ANGERS | LOPEX’93 | |
|---|---|---|---|---|---|
| 400–2500 | mean | 4.51 × 10−4 | 4.52 × 10−4 | 6.18 × 10−3 | 4.65 × 10−3 |
| maximum | 3.18 × 10−3 | 3.54 × 10−3 | 2.86 × 10−2 | 1.87 × 10−2 | |
| 400–680 | mean | 6.99 × 10−5 | 7.78 × 10−5 | 1.10 × 10−3 | 6.36 × 10−4 |
| maximum | 4.48 × 10−4 | 4.83 × 10−4 | 3.35 × 10−3 | 4.19 × 10−3 | |
| 400–800 | mean | 1.12 × 10−4 | 1.19 × 10−4 | 2.91 × 10−3 | 3.38 × 10−3 |
| maximum | 5.81 × 10−4 | 6.32 × 10−4 | 9.56 × 10−3 | 1.77 × 10−2 | |
| 900–2500 | mean | 5.57 × 10−5 | 5.56 × 10−5 | 4.96 × 10−3 | 2.51 × 10−3 |
| maximum | 1.69 × 10−4 | 1.75 × 10−4 | 2.37 × 10−2 | 8.99 × 10−3 | |
| Cab μg/cm2 | Car μg/cm2 | EWT cm | SLWg/cm2 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | S4 | S1 | S2 | S3 | S4 | S1 | S2 | S3 | S4 | S1 | S2 | S3 | S4 | |
| PC1 | 0.06 | 0.81 | 0.18 | 0.00 | 0.04 | 0.42 | 0.10 | 0.00 | 0.38 | 0.02 | 0.08 | 0.49 | 0.55 | 0.06 | 0.21 | 0.59 |
| PC2 | 0.14 | 0.67 | 0.83 | 0.00 | 0.06 | 0.29 | 0.40 | 0.00 | 0.69 | 0.01 | 0.01 | 0.94 | 0.38 | 0.04 | 0.03 | 0.40 |
| PC3 | 0.62 | 0.00 | 0.11 | 0.00 | 0.30 | 0.18 | 0.00 | 0.00 | 0.23 | 0.00 | 0.00 | 0.04 | 0.03 | 0.00 | 0.01 | 0.31 |
| PC4 | 0.11 | 0.16 | 0.02 | 0.00 | 0.05 | 0.09 | 0.18 | 0.00 | 0.14 | 0.00 | 0.00 | 0.14 | 0.01 | 0.01 | 0.00 | 0.07 |
| PC5 | 0.01 | 0.10 | 0.00 | 0.00 | 0.00 | 0.02 | 0.12 | 0.00 | 0.00 | 0.01 | 0.01 | 0.01 | 0.36 | 0.02 | 0.01 | 0.30 |
| PC6 | 0.06 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.12 | 0.00 | 0.01 | 0.00 | 0.01 | 0.00 | 0.00 | 0.01 | 0.02 | 0.01 |
| PC7 | 0.01 | 0.01 | 0.00 | 0.00 | 0.17 | 0.00 | 0.07 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.01 | 0.01 | 0.07 |
| PC8 | 0.00 | 0.01 | 0.00 | 0.00 | 0.01 | 0.00 | 0.06 | 0.00 | 0.03 | 0.00 | 0.04 | 0.01 | 0.01 | 0.00 | 0.11 | 0.02 |
| PC9 | 0.02 | 0.01 | 0.03 | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 | 0.01 | 0.04 | 0.05 | 0.00 | 0.21 | 0.10 | 0.14 | 0.00 |
| PC10 | 0.00 | 0.00 | 0.01 | 0.00 | 0.06 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.10 | 0.00 | 0.02 | 0.00 | 0.22 | 0.00 |
| Spectral Index | Regression Model | R2 |
|---|---|---|
| NDVI | (Cab) | 0.78 |
| (Car) | 0.51 | |
| RVI | (Cab) | 0.89 |
| (Car) | 0.59 | |
| MTCI | (Cab) | 0.90 |
| y = (Car) | 0.43 | |
| ND705 | (Cab) | 0.94 |
| y = (Car) | 0.61 | |
| PRI | (Cab) | 0.02 |
| y = (Car) | 0.21 | |
| SIPI | (Cab) | 0.72 |
| y = (Car) | 0.41 | |
| CIgreen | (Cab) | 0.91 |
| (Car) | 0.53 | |
| CIred edge | (Cab) | 0.89 |
| (Car) | 0.50 | |
| NDWI | y = 0.58x − 0.01 (EWT) | 0.95 |
| WI | y = 0.68x − 0.69 (EWT) | 0.95 |
| MSI | (EWT) | 0.97 |
| NDLMA | y = (SLW) | 0.83 |
| NDMI | y = (SLW) | 0.97 |
| Simulated | ANGERS | LOPEX’93 | |||
|---|---|---|---|---|---|
| SLW | NDLMA | R2 | 0.78 | 0.79 | 0.56 |
| RMSE | 0.0028 | 0.0018 | 0.0018 | ||
| NDMI | R2 | 0.95 | 0.73 | 0.7 | |
| RMSE | 0.0016 | 0.0025 | 0.0021 | ||
| PCA | R2 | 0.92 | 0.81 | 0.58 | |
| RMSE | 0.00169 | 0.00173 | 0.00178 | ||
| EWT | NDWI | R2 | 0.95 | 0.56 | 0.53 |
| RMSE | 0.0041 | 0.0044 | 0.0073 | ||
| WI | R2 | 0.96 | 0.75 | 0.61 | |
| RMSE | 0.0053 | 0.004 | 0.0094 | ||
| MSI | R2 | 0.96 | 0.77 | 0.83 | |
| RMSE | 0.0043 | 0.0037 | 0.0028 | ||
| PCA | R2 | 0.99 | 0.83 | 0.85 | |
| RMSE | 0.0018 | 0.0036 | 0.0028 | ||
| Cab (µg/cm2) | Car (µg/cm2) | ||||||
|---|---|---|---|---|---|---|---|
| Simulated | ANGERS | LOPEX’93 | Simulated | ANGERS | LOPEX’93 | ||
| NDVI | R2 | 0.66 | 0.41 | 0.02 | 0.26 | 0.35 | 0.003 |
| RMSE | 17.74 | 18.02 | 27.9 | 9.42 | 4.09 | 6.67 | |
| RVI | R2 | 0.75 | 0.14 | 0.004 | 0.29 | 0.12 | 0.0001 |
| RMSE | 14.31 | 26.4 | 34.08 | 9.03 | 6.19 | 7.93 | |
| MTCI | R2 | 0.9 | 0.59 | 0.13 | 0.22 | 0.64 | 0.04 |
| RMSE | 8.89 | 18.23 | 27.59 | 10.05 | 3.41 | 6.37 | |
| ND705 | R2 | 0.89 | 0.95 | 0.12 | 0.38 | 0.84 | 0.07 |
| RMSE | 10.09 | 5.41 | 25.33 | 8.4 | 2.32 | 6.08 | |
| CIgerrn | R2 | 0.91 | 0.92 | 0.13 | 0.36 | 0.75 | 0.1 |
| RMSE | 8.59 | 7.7 | 24.92 | 8.66 | 3.3 | 6.51 | |
| CIred edge | R2 | 0.89 | 0.95 | 0.14 | 0.3 | 0.71 | 0.07 |
| RMSE | 9.42 | 6.23 | 25.05 | 9.23 | 3.11 | 6.07 | |
| PRI | R2 | 0.02 | 0.33 | 0.01 | 0.35 | 0.18 | 0.0003 |
| RMSE | 29.52 | 23.17 | 26.46 | 9.06 | 6.53 | 6.35 | |
| SIPI | R2 | 0.92 | 0.92 | 0.14 | 0.48 | 0.7 | 0.04 |
| RMSE | 7.91 | 7.39 | 24.52 | 7.77 | 4.63 | 7.34 | |
| PCA | R2 | 0.92 | 0.92 | 0.14 | 0.73 | 0.71 | 0.13 |
| RMSE | 7.88 | 6.34 | 24.51 | 5.96 | 2.77 | 5.76 | |
| Leaf Biochemistries | Method | ANGERS | LOPEX’93 |
|---|---|---|---|
| EWT cm | PCA | 0.0036 | 0.0028 |
| P-5 | 0.0020 | 0.0017 | |
| SLW g/cm2 | PCA | 0.00173 | 0.00178 |
| P-5 | 0.0026 | 0.0034 | |
| Cab µg/cm2 | PCA | 6.34 | 24.51 |
| P-5 | 5.17 | 32.35 | |
| Car µg/cm2 | PCA | 2.41 | 6.20 |
| P-5 | 2.77 | 5.76 |
| Simulated Validation | ANGERS | LOPEX | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R² | RMSE | Bias | SL | INTC | R² | RMSE | Bias | SL | INTC | R² | RMSE | Bias | SL | INTC | |
| SLW | 0.92 | 0.00168 | −2.0% | 0.9 | 7 × 10−4 | 0.81 | 0.00173 | 29.8% | 0.656 | 2 × 10−4 | 0.58 | 0.00178 | 3.6% | 0.845 | 8 × 10−4 |
| EWT | 0.99 | 0.002 | −1.3% | 0.984 | 3 × 10−4 | 0.83 | 0.0036 | 9.7% | 1.40 | −0 | 0.85 | 0.0028 | 9.8% | 1.09 | 0 |
| Cab | 0.92 | 7.88 | −1.3% | 0.903 | 4.61 | 0.92 | 6.34 | 11.2% | 0.847 | 8.98 | 0.14 | 24.51 | 3.1% | 0.182 | 42.08 |
| Car | 0.73 | 5.96 | −7.6% | 0.54 | 5.00 | 0.71 | 2.77 | 1.9% | 0.759 | 2.24 | 0.13 | 5.76 | 11.6% | 0.2 | 9.93 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Song, B.; Zhang, S.; Liu, X. A Novel Principal Component Analysis Method for the Reconstruction of Leaf Reflectance Spectra and Retrieval of Leaf Biochemical Contents. Remote Sens. 2017, 9, 1113. https://doi.org/10.3390/rs9111113
Liu L, Song B, Zhang S, Liu X. A Novel Principal Component Analysis Method for the Reconstruction of Leaf Reflectance Spectra and Retrieval of Leaf Biochemical Contents. Remote Sensing. 2017; 9(11):1113. https://doi.org/10.3390/rs9111113
Chicago/Turabian StyleLiu, Liangyun, Bowen Song, Su Zhang, and Xinjie Liu. 2017. "A Novel Principal Component Analysis Method for the Reconstruction of Leaf Reflectance Spectra and Retrieval of Leaf Biochemical Contents" Remote Sensing 9, no. 11: 1113. https://doi.org/10.3390/rs9111113