First Insights into the Gut Microbiota of Mexican Patients with Celiac Disease and Non-Celiac Gluten Sensitivity
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Considerations
2.2. Recruitment of Participants
2.3. DNA Extraction, PCR, and 16S rDNA Sequencing
2.4. Bioinformatics
2.5. Statistical Analysis
3. Results
3.1. Subjects
3.2. 16S Sequencing and Taxonomic Classification of Sequence Reads
3.3. Microbiota at Baseline
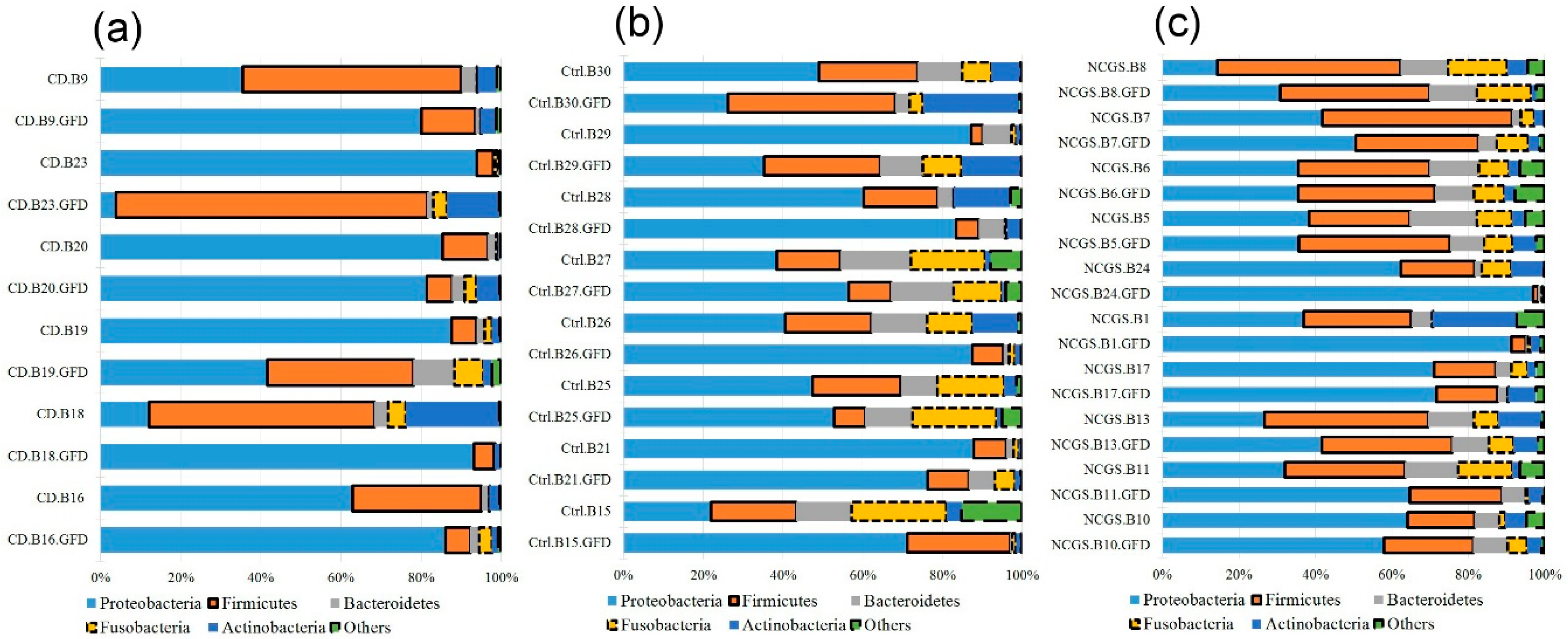
3.3.1. Microbiota in Duodenal Biopsy Samples at Baseline
3.3.2. Microbiota in Fecal Samples at the Baseline
3.4. Effect of GFD on the Gut Microbiota
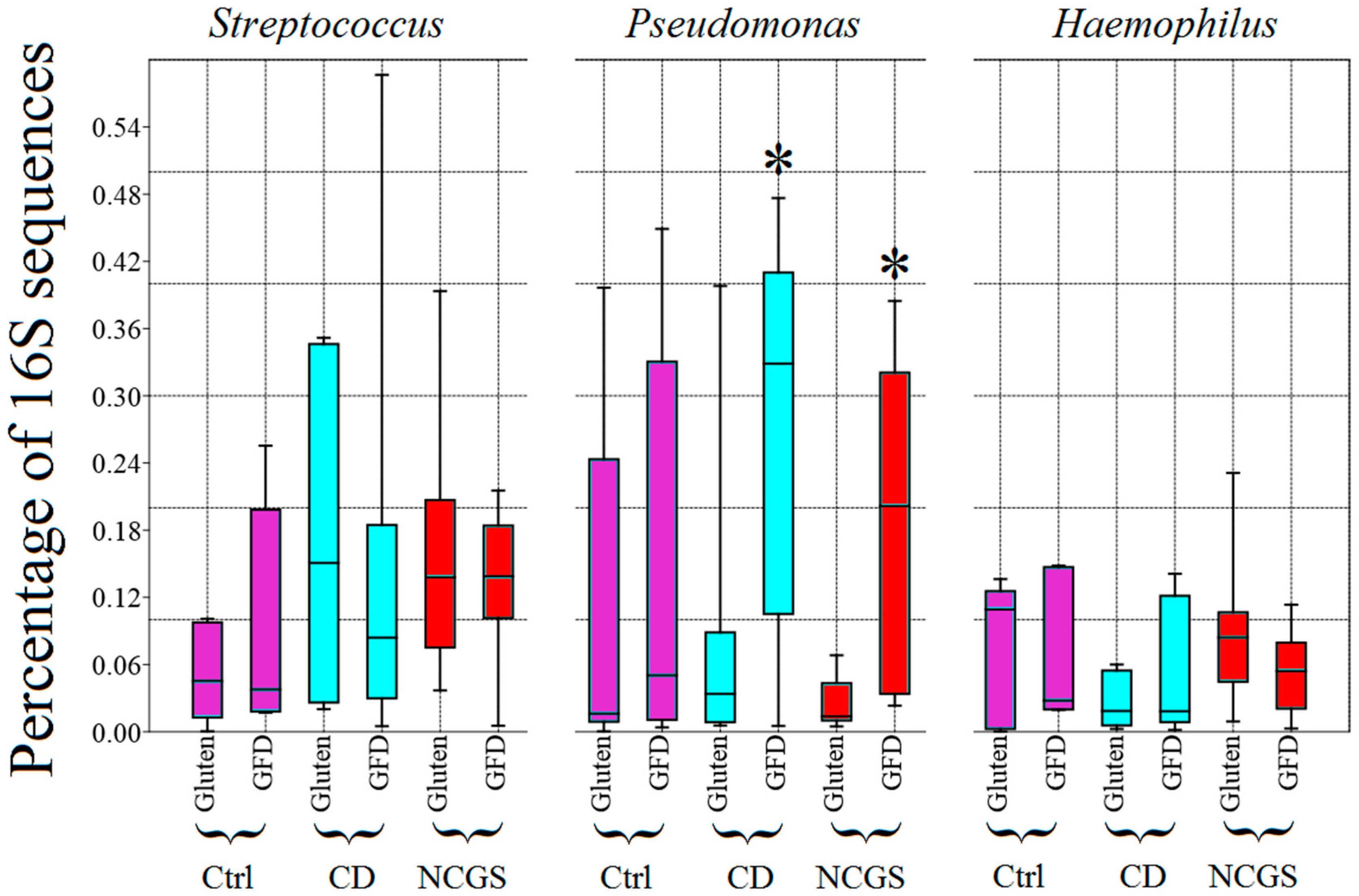
3.4.1. Effect of GFD on Duodenal Microbiota
3.4.2. Effect of GFD on Fecal Microbiota
3.4.3. Effect of GFD on the Predicted Functional Profile
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Inohara, N. Regulation of the gut microbiota by the mucosal immune system in mice. Int. Immunol. 2014, 26, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Walters, W.A.; Xu, Z.; Knight, R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014, 588, 4223–4233. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Chang, E.B. Exploring gut microbes in human health and disease: Pushing the envelope. Genes Dis. 2014, 1, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Ludvigsson, J.F.; Leffler, D.A.; Bai, J.; Biagi, F.; Fasano, A.; Green, P.H.; Hadjivassiliou, M.; Kaukinen, K.; Kelly, C.P.; Leonard, J.N.; et al. The Oslo definitions for coeliac disease and related terms. Gut 2013, 62, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Galipeau, H.J.; Verdu, E.F. Gut microbes and adverse food reactions: Focus on gluten related disorders. Gut Microbes 2014, 5, 594–605. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Achury, J.; Zhernakova, A.; Pulit, S.L.; Trynka, G.; Hunt, K.A.; Romanos, J.; Raychaudhuri, S.; van Heel, D.A.; Wijmenga, C.; de Bakker, P.I. Fine mapping in the MHC region accounts for 18% additional genetic risk for celiac disease. Nat. Genet. 2015, 47, 577–578. [Google Scholar] [CrossRef] [PubMed]
- Escudero-Hernandez, C.; Peña, A.S.; Bernardo, D. Immunogenetic pathogenesis of celiac disease and non-celiac gluten sensitivity. Curr. Gastroenterol. Rep. 2016, 18, 36. [Google Scholar] [CrossRef] [PubMed]
- Rotondi Aufiero, V.; Fasano, A.; Mazzarella, G. Non-celiac gluten sensitivity: How its gut immune activation and potential dietary management differ from celiac disease. Mol. Nutr. Food. Res. 2018, 62, e1700854. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Elli, L.; Bonaz, B.; Bouma, G.; Carroccio, A.; Castillejo, G.; Cellier, C.; Cristofori, F.; de Magistris, L.; Dolinsek, J.; et al. Diagnosis of non-celiac gluten sensitivity (NCGS): The Salerno experts’ criteria. Nutrients 2015, 7, 4966–4977. [Google Scholar] [CrossRef] [PubMed]
- Verdu, E.F.; Galipeau, H.J.; Jabri, B. Novel players in coeliac disease pathogenesis: Role of the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Ercolini, D.; Francavilla, R.; Vannini, L.; De Filippis, F.; Capriati, T.; Di Cagno, R.; Iacono, G.; De Angelis, M.; Gobbetti, M. From an imbalance to a new imbalance: Italian-style gluten-free diet alters the salivary microbiota and metabolome of African celiac children. Sci. Rep. 2015, 5, 18571. [Google Scholar] [CrossRef] [PubMed]
- Verdu, E.F.; Armstrong, D.; Murray, J.A. Between celiac disease and irritable bowel syndrome: The “no man’s land” of gluten sensitivity. Am. J. Gastroenterol. 2009, 104, 1587–1594. [Google Scholar] [CrossRef] [PubMed]
- Volta, U.; Caio, G.; Tovoli, F.; De Giorgio, R. Non-celiac gluten sensitivity: Questions still to be answered despite increasing awareness. Cell. Mol. Immunol. 2013, 10, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Rausch, P.; Wang, J.; Skieceviciene, J.; Kiudelis, G.; Bhagalia, K.; Amarapurkar, D.; Kupcinskas, L.; Schreiber, S.; Rosenstiel, P.; et al. Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut 2015, 65, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mazcorro, J.F.; Noratto, G.; Remes-Troche, J.M. The effect of gluten-free diet on health and the gut microbiota cannot be extrapolated from one population to others. Nutrients 2018, 10, 1421. [Google Scholar] [CrossRef]
- Golfetto, L.; de Senna, F.D.; Hermes, J.; Soares Beserra, B.T.; da Silva França, F.; Martinello, F. Lower bidifobacteria counts in adult patients with celiac disease on a gluten-free diet. Arg. Gastroenterol. 2014, 51, 139–143. [Google Scholar] [CrossRef]
- Cheng, J.; Kalliomäki, M.; Heilig, H.G.H.J.; Palva, A.; Lähteenoja, H.; de Vos, W.M.; Salojärvi, J.; Satokari, R. Duodenal microbiota composition and mucosal homeostasis in pediatric celiac disease. BMC Gastroenterol. 2013, 13, 113. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, E.; Donat, E.; Ribes-Koninckx, C.; Fernandez-Murga, M.L.; Sanz, Y. Duodenal-mucosal bacteria associated with celiac disease in children. Appl. Environ. Microbiol. 2013, 79, 5472–5479. [Google Scholar] [CrossRef] [PubMed]
- Bonder, M.J.; Tigchelaar, E.F.; Cai, X.; Trynka, G.; Cenit, M.C.; Hrdlickova, B.; Zhong, H.; Vatanen, T.; Gevers, D.; Wijmenga, C.; et al. The influence of a short-term gluten free diet on the human gut microbiota. Genome Med. 2016, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Remes-Troche, J.M. Doctor, why am I celiac if I’m Mexican? Breaking another paradigm of celiac disease in Mexico. Rev. Gastroenterol. Mex. 2018, 83, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Cervantes, K.L.; Remes-Troche, J.M.; Milke-García, M.P.; Romero, V.; Uscanga, L.F. Characteristics and factors related to quality of life in Mexican mestizo patients with celiac disease. BMC Gastroenterol. 2015, 15, 4. [Google Scholar] [CrossRef] [PubMed]
- Remes-Troche, J.M.; Nuñez-Alvares, C.; Uscanga-Dominguez, L.F. Celiac disease in Mexican population: An update. Am. J. Gastroenterol. 2013, 108, 283–284. [Google Scholar] [CrossRef] [PubMed]
- Sotelo Cruz, N.; Calderon de la Barca, A.M.; Hurtado Valenzuela, J.G. Celiac disease in children from the northwest of Mexico: Clinical characteristics of 24 cases. Rev. Gastroenterol. Mex. 2013, 78, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Cerda-Contreras, E.; Duarte-Rojo, A.; Granados, J.; Vargas, F.; Uscanga-Dominguez, L.F. Frecuencia de antígenos de histocompatibilidad DQ2 DQ8 en sujetos con diarrea crónica y enfermedad celiaca (EC). Rev. Gastroentol. Mex. 2008, 73 (Suppl. 2), 131–132. [Google Scholar]
- Remes-Troche, J.M.; Uscanga-Domínguez, L.F.; Aceves-Tavares, R.G.; Calderón de la Barca, A.M.; Carmona-Sánchez, R.I.; Cerda-Contreras, E.; Coss-Adame, E.; Icaza-Chávez, M.E.; Lopez-Colombo, A.; Milke-García, M.P.; et al. Clinical guidelines on the diagnosis and treatment of celiac disease in Mexico. Rev. Gastroenterol. Mex. 2018. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mazcorro, J.F.; Lage, N.N.; Mertens-Talcott, S.; Talcott, S.; Chew, B.; Dowd, S.E.; Kawas, J.R.; Noratto, G.D. Effect of dark sweet cherry powder consumption on the gut microbiota, short-chain fatty acids, and biomarkers of gut health in obese db/db mice. PeerJ 2018, 6, e4195. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Meth. 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Rideout, J.R.; He, Y.; Navas-Molina, J.A.; Walters, W.A.; Ursell, L.K.; Gibbons, S.M.; Chase, J.; McDonald, D.; Gonzalez, A.; Robbins-Pianka, A.; et al. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2014, 2, e545. [Google Scholar] [CrossRef] [PubMed]
- Navas-Molina, J.A.; Peralta-Sanchez, J.M.; Gonzalez, A.; McMurdie, P.J.; Vazquez-Baeza, Y.; Xu, Z.; Ursell, L.K.; Lauber, C.; Zhou, H.; Song, S.J.; et al. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol. 2013, 531, 371–444. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–823. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Wladron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mazcorro, J.F.; Mills, D.A.; Murphy, K.; Noratto, G. Effect of barley supplementation on the fecal microbiota, caecal biochemistry, and key biomarkers of obesity and inflammation in obese db/db mice. Eur. J. Nutr. 2017. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Beiko, R.G. Identifying biologically relevant differences between metagenomic communities. Bioinformatics 2010, 26, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Jousset, A.; Bienhold, C.; Chatzinotas, A.; Gallien, L.; Gobet, A.; Kurm, V.; Küsel, K.; Rillig, M.C.; Rivett, D.W.; Salles, J.F.; et al. Where less may be more: How the rare biosphere pulls ecosystems strings. ISME J. 2017, 11, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Jaspers, A.; Overmann, J. Ecological significance of microdiversity: Identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl. Environ. Microbiol. 2004, 70, 4831–4839. [Google Scholar] [CrossRef] [PubMed]
- Caminero, A.; Herrán, A.R.; Nistal, E.; Pérez-Andrés, J.; Vaquero, L.; Vivas, S.; Ruiz de Morales, J.M.; Albillos, S.M.; Casqueiro, J. Diversity of the cultivable human gut microbiome involved in gluten metabolism: Isolation of microorganisms with potential interest for coeliac disease. FEMS Microbiol. Ecol. 2014, 88, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Stams, A.J.M.; de Vos, W.M.; Sánchez-Andrea, I. Enrichment of sulfidogenic bacteria from the human intestinal tract. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [PubMed]
- Caminero, A.; Nistal, E.; Herrán, A.R.; Pérez-Andrés, J.; Vaquero, L.; Vivas, S.; Ruíz de Morales, J.; Casqueiro, J. Gluten Metabolism in Humans. Involvement of the Gut Microbiota. In Wheat and Rice in Disease Prevention and Health, 1st ed.; Watson, R.R., Preedy, V.R., Zibadi, S., Eds.; Elsevier Inc.: Atlanta, GA, USA, 2014; pp. 157–170. [Google Scholar]
- Modi, S.R.; Collins, J.J.; Relman, D.A. Antibiotics and the gut microbiota. J. Clin. Investig. 2014, 124, 4212–4218. [Google Scholar] [CrossRef] [PubMed]
- Tjellström, B.; Högberg, L.; Stenhammar, L.; Fälth-Magnusson, K.; Magnusson, K.-E.; Norin, E.; Sundqvist, T.; Midtvedt, T. Faecal short-chain fatty acid pattern in childhood coeliac disease is normalized after more than one year’s gluten-free diet. Microb. Ecol. Health Dis. 2013, 24, 20905. [Google Scholar] [CrossRef]
- Wacklin, P.; Laurikka, P.; Lindfors, K.; Collin, P.; Salmi, T.; Lähdeaho, M.L.; Saavalainen, P.; Mäki, M.; Mättö, J.; Kurppa, K.; et al. Altered duodenal microbiota composition in celiac disease patients suffering from persistent symptoms on a long-term gluten-free diet. Am. J. Gastroenterol. 2014, 109, 1933–1941. [Google Scholar] [CrossRef] [PubMed]
- Silby, M.W.; Winstanley, C.; Godfrey, S.A.C.; Levy, S.B.; Jackson, R.W. Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 2011, 35, 652–680. [Google Scholar] [CrossRef] [PubMed]
- Aujoulat, F.; Roudière, L.; Picaud, J.C.; Jacquot, A.; Filleron, A.; Neveu, D.; Baum, T.P.; Marchandin, H.; Jumas-Bilak, E. Temporal dynamics of the very premature infant gut dominant microbiota. BMC Microbiol. 2014, 14, 325. [Google Scholar] [CrossRef] [PubMed]
- Mao, B.; Li, D.; Zhao, J.; Liu, X.; Gu, Z.; Chen, Y.Q.; Zhang, H.; Chen, W. In vitro fermentation of lactulose by human gut bacteria. J. Agric. Food Chem. 2014, 62, 10970–10977. [Google Scholar] [CrossRef] [PubMed]
- Petriz, B.A.; Castro, A.P.; Almeida, J.A.; Gomes, C.P.; Fernandes, G.R.; Kruger, R.H.; Pereira, R.W.; Franco, O.L. Exercise induction of gut microbiota modifications in obese, non-obese and hypertensive rats. BMC Genom. 2014, 15, 511. [Google Scholar] [CrossRef] [PubMed]
- Scales, B.S.; Dickson, R.P.; LiPuma, J.J.; Huffnagle, G.B. Microbiology, genomics, and clinical significance of the Pseudomonas fluorescens species complex, an unappreciated colonizer of humans. Clin. Microbiol. 2014, 27, 927–948. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 2015, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Tian, N.; Valery, A.C.; Zhong, Y.; Schuppan, D.; Helmerhorst, E.J. Identification of pseudolysin (lasB) as an aciduric gluten-degrading enzyme with high therapeutic potential for celiac disease. Am. J. Gastroenterol. 2015, 110, 899–908. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, V.; Casaburi, G.; Precone, V.; Pagliuca, C.; Colicchio, R.; Sarnataro, D.; Discepolo, V.; Kim, S.M.; Russo, I.; Del Vecchio Blanco, G.; et al. Metagenomics reveals dysbiosis and a potentially pathogenic N. flavescens strain in duodenum of adult celiac patients. Am. J. Gastroenterol. 2016, 111, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell. Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Haberman, Y.; Tickle, T.L.; Dexheimer, P.J.; Kim, M.O.; Tang, D.; Karns, R. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J. Clin. Investig. 2014, 124, 3617–3633. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Ghoshal, U.; Dhole, T.N.; Ghoshal, U.C. Fecal microbiota in patients with irritable bowel syndrome compared to healthy controls using real-time polymerase chain reaction: An evidence of dysbiosis. Dig. Dis. Sci. 2015, 60, 2953–2962. [Google Scholar] [CrossRef] [PubMed]
- Nistal, E.; Caminero, A.; Vivas, S.; Ruiz de Morales, J.M.; Saenz de Miera, L.E.; Rodríguez-Aparicio, L.B.; Casqueiro, J. Differences in faecal bacteria populations and faecal bacteria metabolism in healthy adults and celiac disease patients. Biochimie 2012, 94, 1724–1729. [Google Scholar] [CrossRef] [PubMed]
- De Palma, G.; Nadal, I.; Medina, M.; Donat, E.; Ribes-Koninckx, C.; Calabuig, M.; Sanz, Y. Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. 2010, 10, 63. [Google Scholar] [CrossRef] [PubMed]
- Jakobsdottir, G.; Bjerregaard, J.H.; Skovbjerg, H.; Nyman, M. Fasting serum concentration of short-chain fatty acids in subjects with microscopic colitis and celiac disease: No difference compared with controls, but between genders. Scand. J. Gastroenterol. 2013, 48, 696–701. [Google Scholar] [CrossRef] [PubMed]
- Fairweather, D.; Rose, N.R. Women and autoimmune diseases. Emerg. Infect. Dis. 2004, 10, 2005–2011. [Google Scholar] [CrossRef] [PubMed]
- Harnett, J.; Myers, S.P.; Rolfe, M. Significantly higher faecal counts of the yeasts candida and saccharomyces identified in people with coealic disease. Gut Pathog. 2017, 9, 26. [Google Scholar] [CrossRef] [PubMed]







| ID | Health Status | Age | BMI | Sex | Paired Duodenal Samples? | Paired Fecal Samples? |
|---|---|---|---|---|---|---|
| 9 | CD | 35 | 27 | Woman | Yes | Yes |
| 16 | CD | 36 | 20 | Woman | Yes | Yes |
| 18 | CD | 62 | 18 | Woman | Yes | Yes |
| 19 | CD | 25 | 23 | Woman | Yes | NA |
| 20 | CD | 47 | 25 | Woman | Yes | NA |
| 23 | CD | 73 | 21 | Woman | Yes | Only on GFD |
| 1 | NCGS | 23 | 28 | Woman | Yes | Yes |
| 3 | NCGS | 21 | 24 | Woman | Only baseline | NA |
| 5 | NCGS | 24 | 25 | Woman | Yes | Only on GFD |
| 6 | NCGS | 23 | 29 | Woman | Yes | Yes |
| 7 | NCGS | 22 | 25 | Woman | Yes | NA |
| 8 | NCGS | 24 | 27 | Woman | Yes | NA |
| 10 | NCGS | 27 | 23 | Man | Yes | Yes |
| 11 | NCGS | 23 | 29 | Man | Yes | Only baseline |
| 13 | NCGS | 37 | 31 | Woman | Yes | Yes |
| 17 | NCGS | 59 | 19 | Woman | Yes | Yes |
| 22 | NCGS | 34 | 26 | Woman | Only baseline | NA |
| 24 | NCGS | 38 | 24 | Woman | Yes | Only baseline |
| 2 | Control | 23 | 33 | Man | Only baseline | NA |
| 4 | Control | 24 | 33 | Man | Only baseline | NA |
| 12 | Control | 23 | 24 | Woman | Only baseline | NA |
| 14 | Control | 25 | 23 | Woman | Only baseline | NA |
| 15 | Control | 26 | 21 | Woman | Yes | Yes |
| 21 | Control | 24 | 29 | Man | Yes | NA |
| 25 | Control | 45 | 28 | Woman | Yes | Only baseline |
| 26 | Control | 64 | 24 | Man | Yes | Yes |
| 27 | Control | 23 | 25 | Man | Yes | Only baseline |
| 28 | Control | 39 | 25 | Man | Yes | NA |
| 29 | Control | 58 | 26 | Woman | Yes | NA |
| 30 | Control | 42 | 27 | Woman | Yes | NA |
| CD (n = 6) | NCGS (n = 12) | Controls (n = 12) | p Value | |
|---|---|---|---|---|
| Proportion of women | 100% | 92% | 50% | 0.017 |
| CD in family, % | 67% | 17% | 8% | 0.017 |
| DQ2 or DQ8 positive, % | 83% | 50% | 42% | 0.217 |
| Severe abdominal bloating (Likert), % | 66% | 81% | NA | 0.121 |
| Severe abdominal pain (Likert), % | 50% | 42% | NA | 0.862 |
| Age in years (median, range) | 41.5 (25–73) | 24 (21–59) | 25.5 (23–64) | 0.077 |
| BMI, kg/m2, median (range) | 21.8 (18–27) | 25.3 (21–30) | 25.2 (19–31) | 0.050 |
| Hemoglobin, g/dL, median (range) | 12.3 (10.7–12.6) | 13.8 (12.1–14.6) | 13.8 (12.7–16) | 0.050 |
| Total cholesterol, mg/dL, median (range) | 151 (110–222) | 207 (116–323) | 198 (136–299) | 0.100 |
| HDL, mg/dL, median (range) | 38 (35–47) | 43 (29–51) | 36 (34–70) | 0.013 |
| LDL, mg/dL, median (range) | 91.8 (63–161) | 109 (75–143) | 106 (79–186) | 0.409 |
| Triglycerides, mg/dL, median (range) | 69.5 (40–230) | 108 (62–270) | 154 (83–277) | 0.182 |
| AST, median (range) UI/mL | 29 (19–37) | 23 (8–44) | 26 (8–53) | 0.523 |
| ALT, median (range) UI/mL | 22 (10–39) | 19 (11–85) | 24 (11–51) | 0.895 |
| Eosinophils DLP, median (range) | 5 (0–22) | 1.5 (0–13) | 3.8 (0–11) | 0.392 |
| IEL in duodenum, median (range) | 24 (15–39) | 8 (0–22) | 6 (0–12) | 0.001 |
| Biopsy Samples | Baseline (n = 30) | On GFD (n = 24) | p Value | Control (n = 20) | CD (n = 12) | NCGS (n = 22) | p Value |
|---|---|---|---|---|---|---|---|
| Richness | 1127 | 1177 | 0.9562 | 1695 | 1687 | 1917 | 0.8450 |
| PD whole tree | 87 | 91 | 0.8961 | 1113 | 1057 | 1231 | 0.8426 |
| Chao1 | 1746 | 1831 | 0.9924 | 88 | 86 | 91 | 0.9327 |
| Shannon | 5.5 | 5.5 | 0.8954 | 5.6 a | 4.8 a,b | 5.9 a,c | 0.0193 |
| Fecal samples | Baseline (n = 14) | On GFD (n = 12) | p value | Control (n = 6) | CD (n = 7) | NCGS (n = 13) | p value |
| Richness | 1692 | 1341 | 0.2519 | 1696 | 1347 | 1552 | 0.7946 |
| PD whole tree | 101 | 89 | 0.3217 | 99 | 86 | 98 | 0.6694 |
| Chao1 | 2205 | 1826 | 0.2519 | 2192 | 1783 | 2088 | 0.8086 |
| Shannon | 5.5 | 5.2 | 0.7425 | 4.6 | 5.8 | 5.4 | 0.7551 |
| CD (n = 6) | NCGS (n = 12) | Controls (n = 12) | ||||
|---|---|---|---|---|---|---|
| Baseline | On GFD | Baseline | On GFD | Baseline | On GFD | |
| Hemoglobin, g/dL, median (range) | 12.3 (10.7–12.6) | 12.4 (12.2–13.3) | 13.8 (12.1–14.6) | 13.5 (11.6–12.10) | 13.8 (12.7–16) | 13.7 (12.7–14.5) |
| Total cholesterol, mg/dL, median (range) | 151 (110–222) | 160.5 (103–210) | 207 (116–323) | 185 (140–245) | 198 (136–299) | 175 (189–207) |
| HDL, mg/dL, median (range) | 38 (35–47) | 37.4 (35–37) | 43 (29–51) | 41 (14–53.5) | 36 (34–70) | 46 (15–65) |
| LDL, mg/dL, median (range) | 91.8 (63–161) | 104 (65–130) | 109 (75–143) | 121 (76–135) | 106 (79–186) | 120 (82–157) |
| Triglycerides, mg/dL, median (range) | 69.5 (40–230) | 118 (38–157) | 108 (62–270) | 93 (70–217) | 154 (83–277) | 194 (73–247) |
| AST, UI/mL, median (range) | 29 (19–37) | 21 (13–40) | 23 (8–44) | 20 (12–52) | 26 (8–53) | 19 (13–28) |
| ALT, UI/mL, median (range) | 22 (10–39) | 25 (16–38) | 19 (11–85) | 22 (9–V61) | 24 (11–51) | 26 (19–33) |
| Severe abdominal bloating (Likert), % | 66% | 0% | 81% | 25% | NA | NA |
| Severe abdominal pain (Likert), % | 50% | 0% | 42% | 14% | NA | NA |
| Eosinophils DLP, median (range) | 5 (0–22) | 10 (0–14) | 1.5 (0–13) | 1.5 (0–15) | 3.8 (0–11) | 4 (0–22) |
| Intraepithelial lymphocytes in duodenum, median (range) | 12 (0–29) | 11 (0–43) | 0 (0–22) | 7.5 (0–22) | 5 (0–20) | 7.5 (5–33) |
| Biopsy Samples | Fecal Samples | |||
| Adonis Test Results | Weighted | Unweighted | Weighted | Unweighted |
| Diet | p = 0.053 | p = 0.293 | p = 0.406 | p = 0.877 |
| Disease | p = 0.072 | p = 0.006 | p = 0.323 | p = 0.195 |
| Group | p = 0.067 | p = 0.080 | p = 0.417 | p = 0.494 |
| Age | p = 0.119 | p = 0.060 | p = 0.299 | p = 0.201 |
| BMI | p = 0.401 | p = 0.082 | p = 0.007 | p = 0.010 |
| Biopsy Samples | Fecal Samples | |||
| ANOSIM Test Results | Weighted | Unweighted | Weighted | Unweighted |
| Diet | R = 0.038 p = 0.106 | R = 0.014 p = 0.262 | R = 0.009 p = 0.333 | R = −0.071 p = 0.952 |
| Disease | R = 0.078 p = 0.026 | R = 0.058 p = 0.058 | R = 0.082 p = 0.136 | R = 0.055 p = 0.221 |
| Group | R = 0.082 p = 0.039 | R = 0.035 p = 0.174 | R = 0.039 p = 0.307 | R = −0.001 p = 0.482 |
| Age | R = 0.083 p = 0.034 | R = 0.081 p = 0.032 | R = 0.009 p = 0.349 | R = 0.009 p = 0.392 |
| BMI | R = 0.034 p = 0.230 | R = −0.050 p = 0.800 | R = 0.132 p = 0.024 | R = 0.155 p = 0.018 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia-Mazcorro, J.F.; Rivera-Gutierrez, X.; Cobos-Quevedo, O.D.J.; Grube-Pagola, P.; Meixueiro-Daza, A.; Hernandez-Flores, K.; Cabrera-Jorge, F.J.; Vivanco-Cid, H.; Dowd, S.E.; Remes-Troche, J.M. First Insights into the Gut Microbiota of Mexican Patients with Celiac Disease and Non-Celiac Gluten Sensitivity. Nutrients 2018, 10, 1641. https://doi.org/10.3390/nu10111641
Garcia-Mazcorro JF, Rivera-Gutierrez X, Cobos-Quevedo ODJ, Grube-Pagola P, Meixueiro-Daza A, Hernandez-Flores K, Cabrera-Jorge FJ, Vivanco-Cid H, Dowd SE, Remes-Troche JM. First Insights into the Gut Microbiota of Mexican Patients with Celiac Disease and Non-Celiac Gluten Sensitivity. Nutrients. 2018; 10(11):1641. https://doi.org/10.3390/nu10111641
Chicago/Turabian StyleGarcia-Mazcorro, Jose F., Xaira Rivera-Gutierrez, Orestes De Jesus Cobos-Quevedo, Peter Grube-Pagola, Arturo Meixueiro-Daza, Karina Hernandez-Flores, Francisco J. Cabrera-Jorge, Hector Vivanco-Cid, Scot E. Dowd, and Jose M. Remes-Troche. 2018. "First Insights into the Gut Microbiota of Mexican Patients with Celiac Disease and Non-Celiac Gluten Sensitivity" Nutrients 10, no. 11: 1641. https://doi.org/10.3390/nu10111641
APA StyleGarcia-Mazcorro, J. F., Rivera-Gutierrez, X., Cobos-Quevedo, O. D. J., Grube-Pagola, P., Meixueiro-Daza, A., Hernandez-Flores, K., Cabrera-Jorge, F. J., Vivanco-Cid, H., Dowd, S. E., & Remes-Troche, J. M. (2018). First Insights into the Gut Microbiota of Mexican Patients with Celiac Disease and Non-Celiac Gluten Sensitivity. Nutrients, 10(11), 1641. https://doi.org/10.3390/nu10111641






