Maternal Consumption of Low-Isoflavone Soy Protein Isolate Confers the Increased Predisposition to Alcoholic Liver Injury in Adult Rat Offspring
Abstract
:1. Introduction
2. Materials and Methods
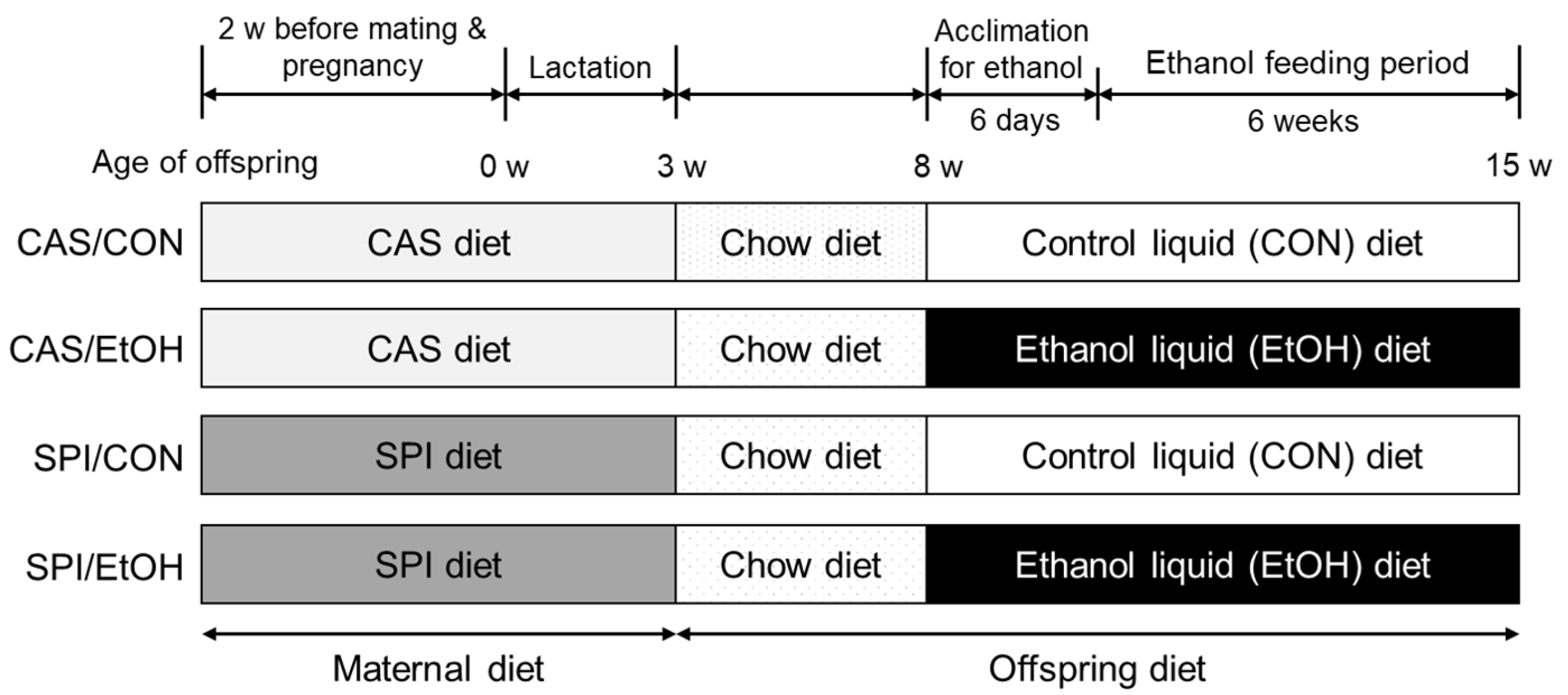
2.1. Animals and Diets
2.2. Serum and Hepatic Biochemical Analysis
2.3. Hepatic SAM and SAH analysis
2.4. Liver Histology
2.5. Total RNA Isolation and Real-Time PCR Analysis
2.6. Semi-Quantitative PCR
2.7. Total Protein Extraction and Immunoblotting
2.8. Statistical Analysis
3. Results
3.1. Effects of Maternal Diet on Body and Organ Weights in Ethanol-Fed Rat Offspring
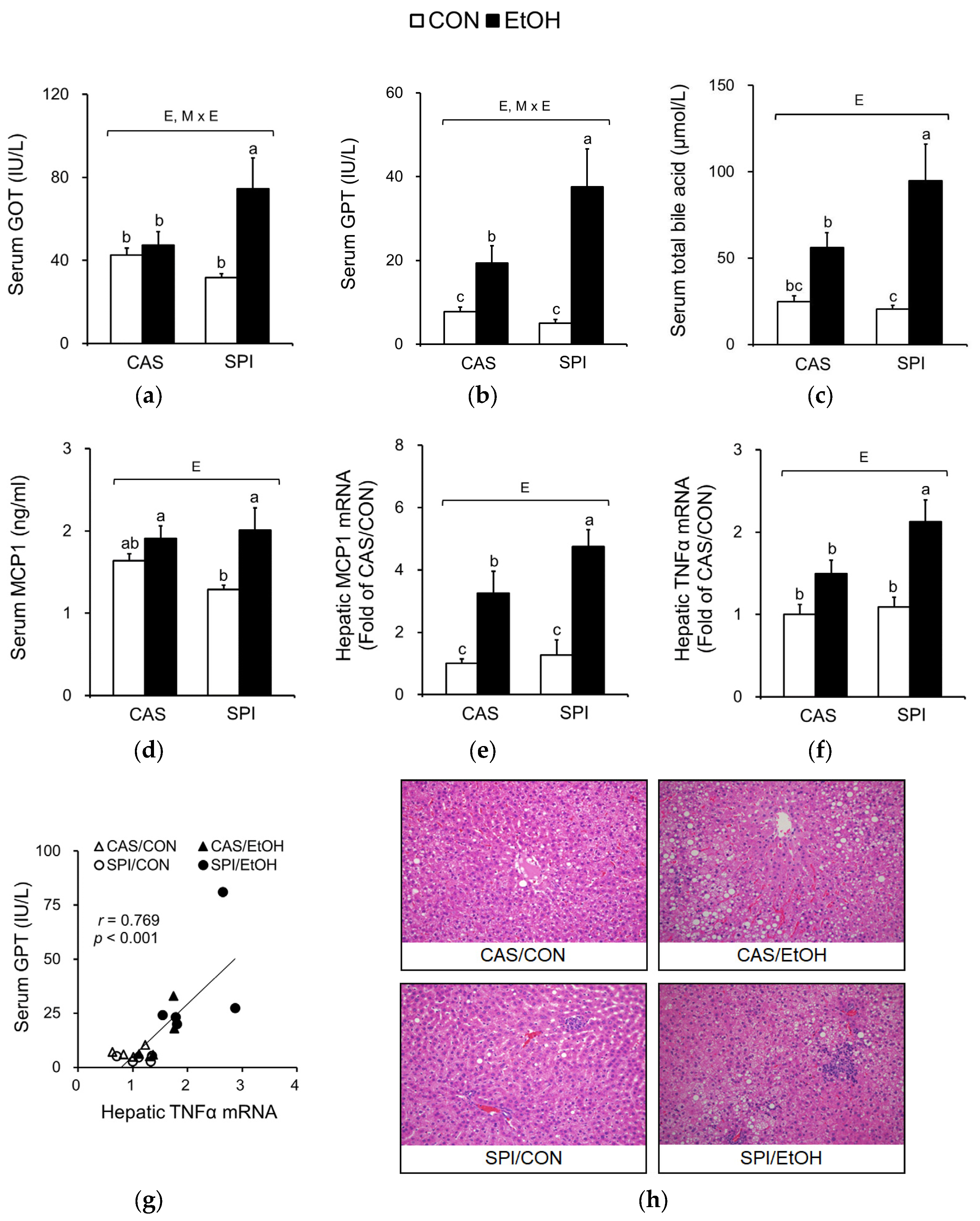
3.2. Effects of Maternal Diet on Liver Injury in Ethanol-Fed Rat Offspring
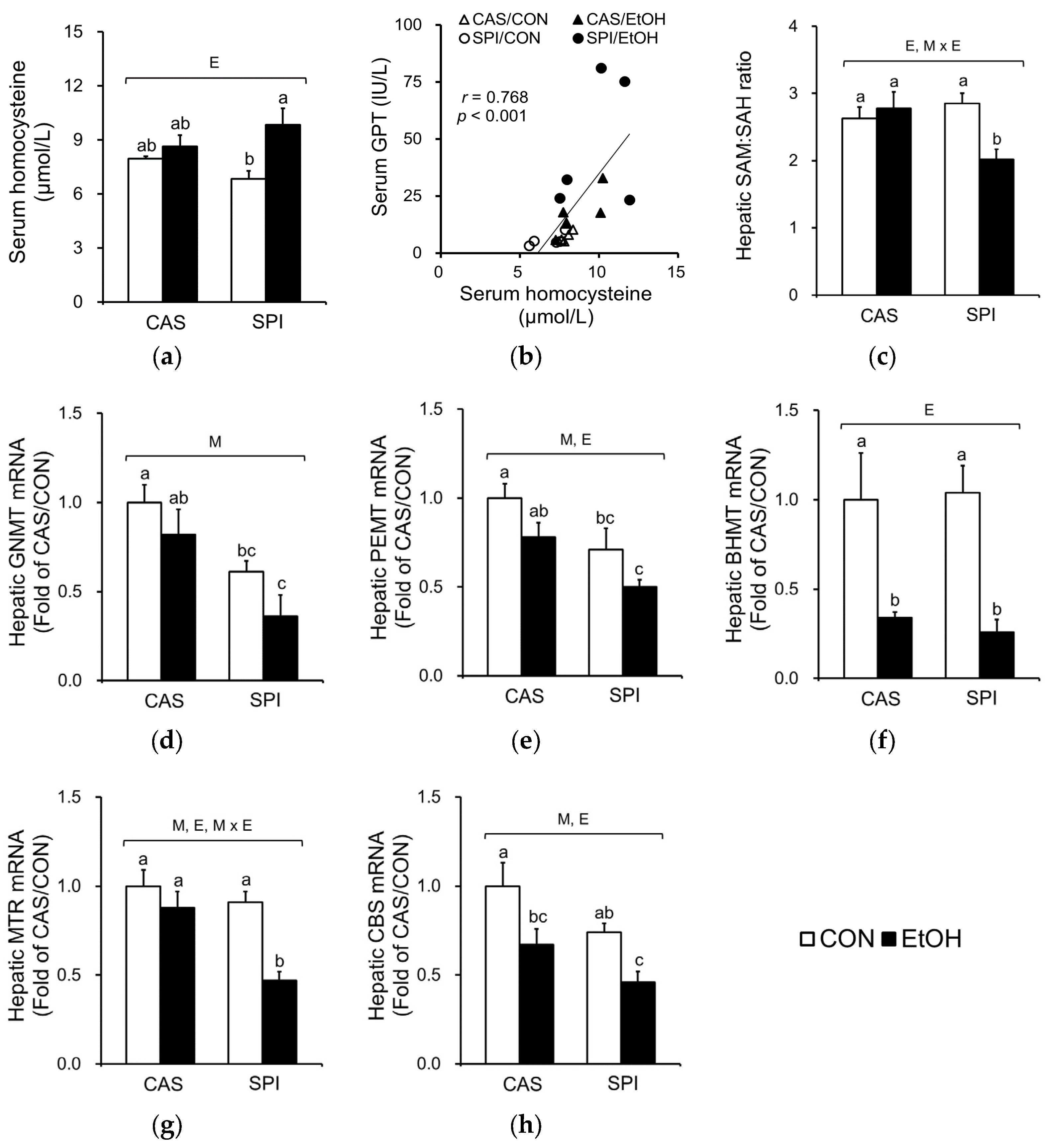
3.3. Effects of Maternal Diet on One-Carbon Metabolism in the Liver of Ethanol-Fed Offspring
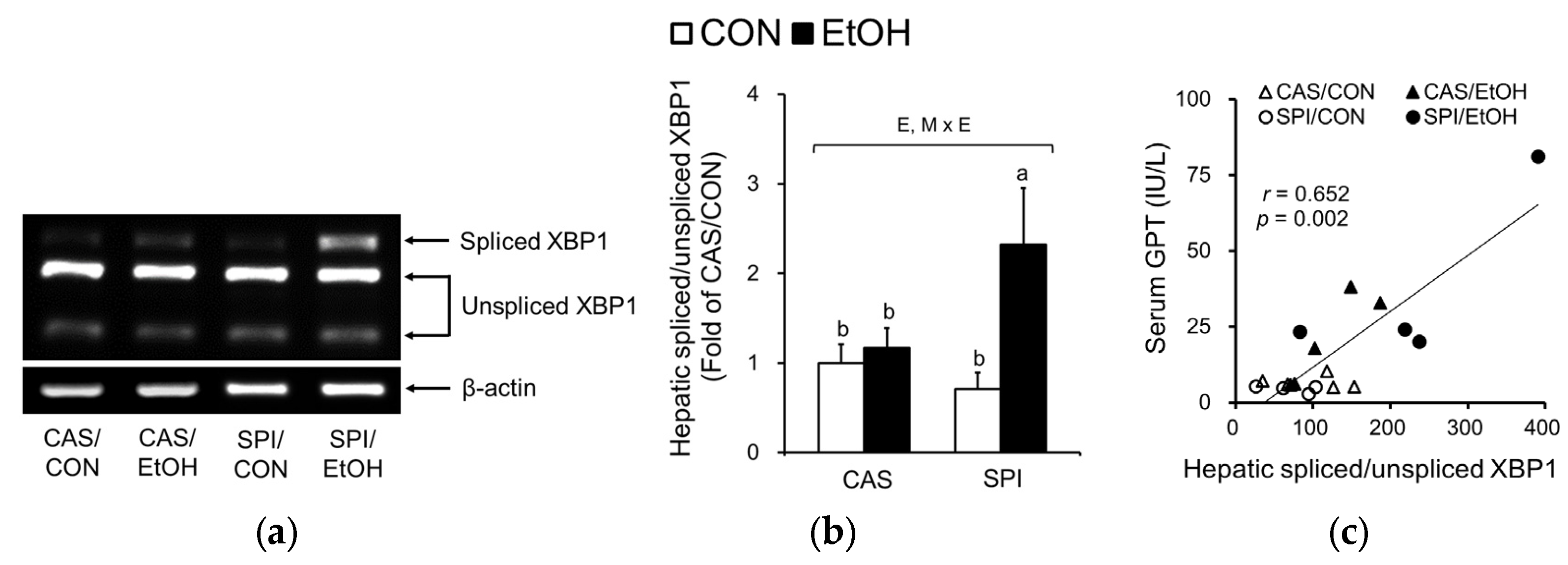
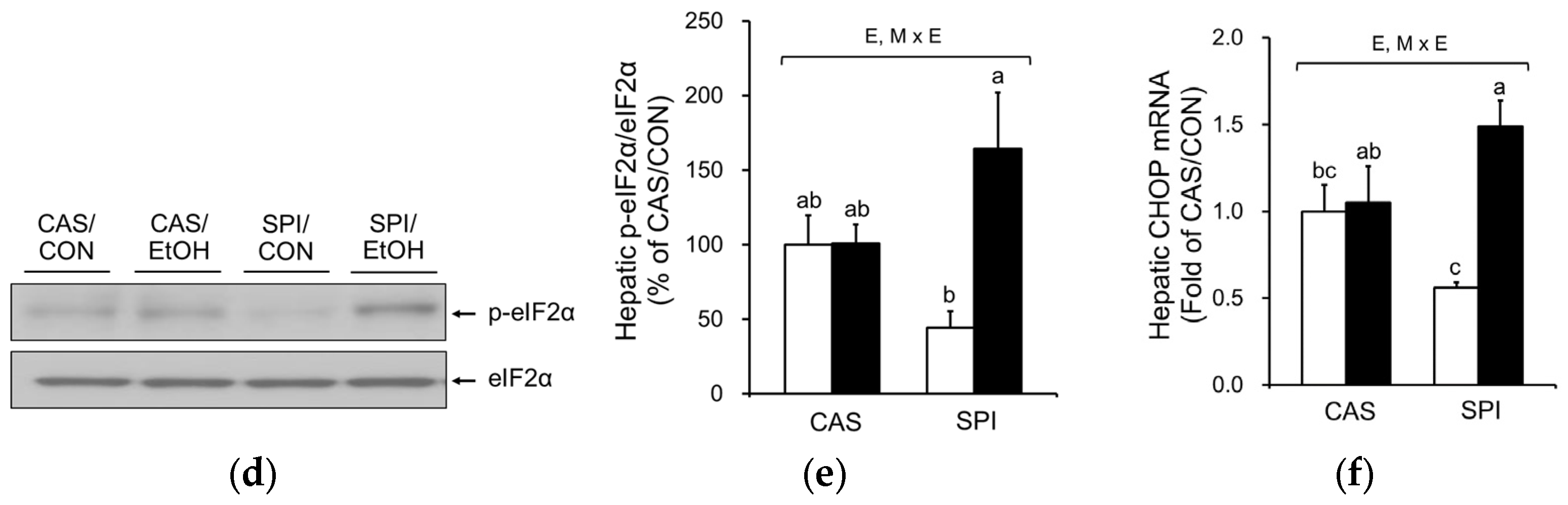
3.4. Effects of Maternal Diet on ER Stress in the Liver of Ethanol-Fed Rat Offspring
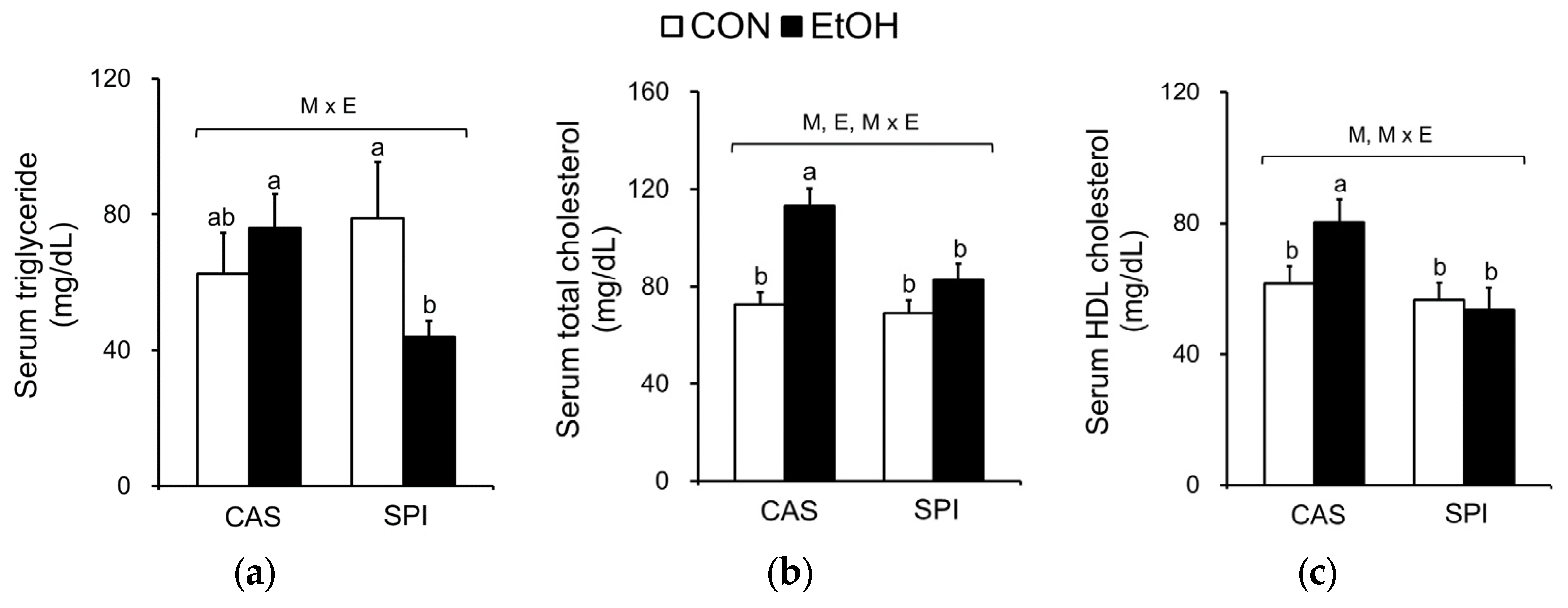
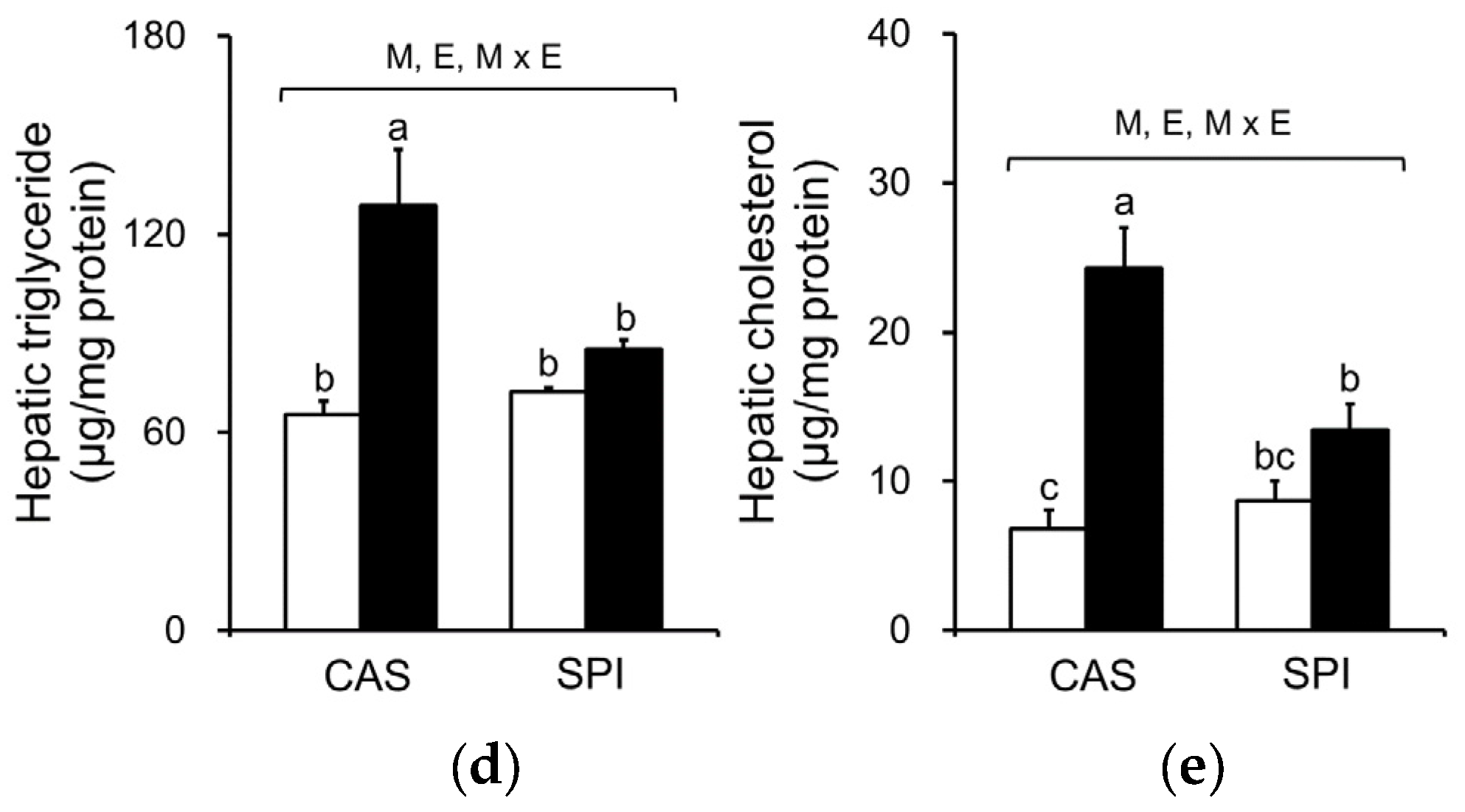
3.5. Effects of Maternal Diet on Serum and Hepatic Lipid Profiles in Ethanol-Fed Rat Offspring
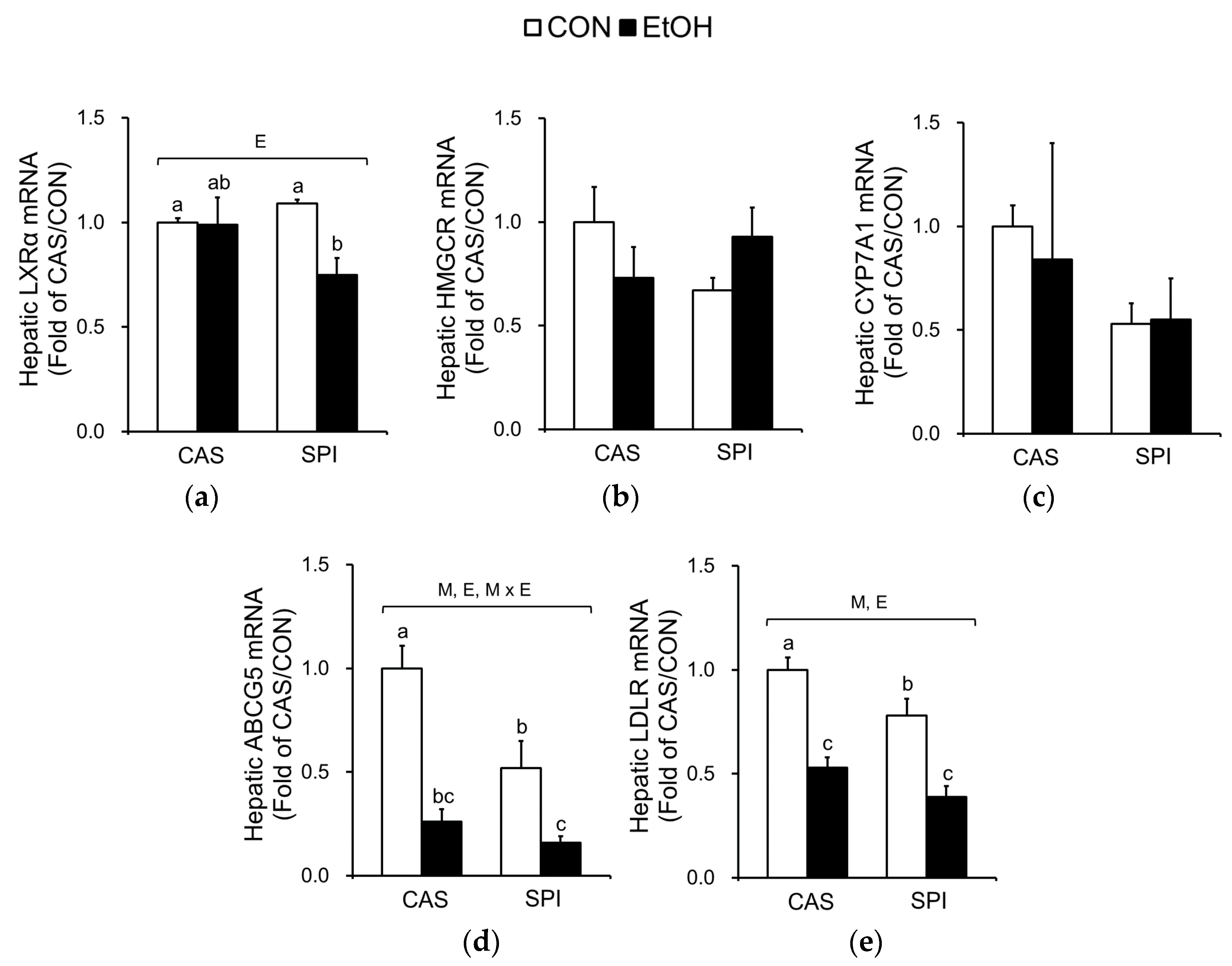
3.6. Effects of Maternal Diet on Serum and Hepatic Cholesterol Metabolism in Ethanol-Fed Rat Offspring
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Martin-Gronert, M.S.; Tarry-Adkins, J.L.; Cripps, R.L.; Chen, J.-H.; Ozanne, S.E. Maternal protein restriction leads to early life alterations in the expression of key molecules involved in the aging process in rat offspring. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 294, R494–R500. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, M.A.; Budge, H.; Symonds, M.E. Early developmental influences on hepatic organogenesis. Organogenesis 2008, 4, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Dudley, K.J.; Sloboda, D.M.; Connor, K.L.; Beltrand, J.; Vickers, M.H. Offspring of mothers fed a high fat diet display hepatic cell cycle inhibition and associated changes in gene expression and DNA methylation. PLoS ONE 2011, 6, e21662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hales, C.N.; Barker, D.J. Type 2 (non-insulin-dependent) diabetes mellitus: The thrifty phenotype hypothesis. Diabetologia 1992, 35, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Navarro, E.; Funtikova, A.N.; Fito, M.; Schroder, H. Prenatal nutrition and the risk of adult obesity: Long-term effects of nutrition on epigenetic mechanisms regulating gene expression. J. Nutr. Biochem. 2017, 39, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kalhan, S.C. One carbon metabolism in pregnancy: Impact on maternal, fetal and neonatal health. Mol. Cell. Endocrinol. 2016, 435, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Won, S.B.; Han, A.; Kwon, Y.H. Maternal consumption of low-isoflavone soy protein isolate alters hepatic gene expression and liver development in rat offspring. J. Nutr. Biochem. 2017, 42, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Han, A.; Won, S.B.; Kwon, Y.H. Different Effects of Maternal Low-Isoflavone Soy Protein and Genistein Consumption on Hepatic Lipid Metabolism of 21-Day-Old Male Rat Offspring. Nutrients 2017, 9, 1039. [Google Scholar] [CrossRef] [PubMed]
- Lieber, C.S. ALCOHOL: Its metabolism and interaction with nutrients. Annu. Rev. Nutr. 2000, 20, 395–430. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, J.A.; Halsted, C.H. Hepatic transmethylation reactions in micropigs with alcoholic liver disease. Hepatology 2004, 39, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Zhou, Z.; Song, M.; Uriarte, S.; Chen, T.; Deaciuc, I.; McClain, C.J. Alcohol-induced S-adenosylhomocysteine accumulation in the liver sensitizes to TNF hepatotoxicity: Possible involvement of mitochondrial S-adenosylmethionine transport. Biochem. Pharmacol. 2007, 74, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Sozio, M.; Crabb, D.W. Alcohol and lipid metabolism. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E10–E16. [Google Scholar] [CrossRef] [PubMed]
- Linz, A.L.; Xiao, R.; Parker, J.G.; Simpson, P.M.; Badger, T.M.; Simmen, F.A. Feeding of soy protein isolate to rats during pregnancy and lactation suppresses formation of aberrant crypt foci in their progeny’s colons: interaction of diet with fetal alcohol exposure. J. Carcinog. 2004, 3, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cahill, L.E.; Peng, C.Y.; Bankovic-Calic, N.; Sankaran, D.; Ogborn, M.R.; Aukema, H.M. Dietary soya protein during pregnancy and lactation in rats with hereditary kidney disease attenuates disease progression in offspring. Br. J. Nutr. 2007, 97, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Reeves, P.G.; Nielsen, F.H.; Fahey, G.C., Jr. AIN-93 purified diets for laboratory rodents: Final report of the american institute of nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 1993, 123, 1939–1951. [Google Scholar] [CrossRef] [PubMed]
- Lieber, C.S.; DeCarli, L.M. Liquid diet technique of ethanol administration: 1989 update. Alcohol Alcohol. 1989, 24, 197–211. [Google Scholar] [PubMed]
- Minniti, G.; Piana, A.; Armani, U.; Cerone, R. Determination of plasma and serum homocysteine by high-performance liquid chromatography with fluorescence detection. J. Chromatogr. A 1998, 828, 401–405. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Stanley, G.H.S. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [PubMed]
- Bottiglieri, T. Isocratic high performance liquid chromatographic analysis of S-adenosylmethionine and S-adenosylhomocysteine in animal tissues: The effect of exposure to nitrous oxide. Biomed. Chromatogr. 1990, 4, 239–241. [Google Scholar] [CrossRef] [PubMed]
- Toda, H.; Suzuki, G.; Nibuya, M.; Shioda, K.; Nishijima, K.; Wakizono, T.; Kanda, Y.; Watanabe, Y.; Shimizu, K.; Nomura, S. Behavioral stress and activated serotonergic neurotransmission induce XBP-1 splicing in the rat brain. Brain Res. 2006, 1112, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Barve, S.; Joshi-Barve, S.; Song, Z.; Hill, D.; Hote, P.; Deaciuc, I.; McClain, C. Interactions of cytokines, S-Adenosylmethionine, and S-Adenosylhomocysteine in alcohol-induced liver disease and immune suppression. J. Gastroenterol. Hepatol. 2006, 21 (Suppl. 3), S38–S42. [Google Scholar] [CrossRef] [PubMed]
- Ji, C. Mechanisms of alcohol-induced endoplasmic reticulum stress and organ injuries. Biochem. Res. Int. 2012, 2012, 216450. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Yang, L.; McCall, S.; Huang, J.; Yu, X.X.; Pandey, S.K.; Bhanot, S.; Monia, B.P.; Li, Y.X.; Diehl, A.M. Inhibiting triglyceride synthesis improves hepatic steatosis but exacerbates liver damage and fibrosis in obese mice with nonalcoholic steatohepatitis. Hepatology 2007, 45, 1366–1374. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yao, T.; Song, Z. Involvement and mechanism of DGAT2 upregulation in the pathogenesis of alcoholic fatty liver disease. J. Lipid Res. 2010, 51, 3158–3165. [Google Scholar] [CrossRef] [PubMed]
- Zannis, V.I.; Chroni, A.; Krieger, M. Role of apoA-I, ABCA1, LCAT, and SR-BI in the biogenesis of HDL. J. Mol. Med. 2006, 84, 276–294. [Google Scholar] [CrossRef] [PubMed]
- Kharbanda, K.K. Role of transmethylation reactions in alcoholic liver disease. World J. Gastroenterol. 2007, 13, 4947–4954. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.P.; Li, Y.S.; Chen, Y.J.; Chiang, E.P.; Li, A.F.; Lee, Y.H.; Tsai, T.F.; Hsiao, M.; Huang, S.F.; Chen, Y.M. Glycine N-methyltransferase−/− mice develop chronic hepatitis and glycogen storage disease in the liver. Hepatology 2007, 46, 1413–1425. [Google Scholar] [CrossRef] [PubMed]
- Tryndyak, V.; de Conti, A.; Kobets, T.; Kutanzi, K.; Koturbash, I.; Han, T.; Fuscoe, J.C.; Latendresse, J.R.; Melnyk, S.; Shymonyak, S.; et al. Interstrain differences in the severity of liver injury induced by a choline- and folate-deficient diet in mice are associated with dysregulation of genes involved in lipid metabolism. FASEB. J. 2012, 26, 4592–4602. [Google Scholar] [CrossRef] [PubMed]
- Oliva, J.; Bardag-Gorce, F.; Li, J.; French, B.A.; French, S.W. S-adenosylmethionine prevents the up regulation of Toll-like receptor (TLR) signaling caused by chronic ethanol feeding in rats. Exp. Mol. Pathol. 2011, 90, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Kharbanda, K.K.; Mailliard, M.E.; Baldwin, C.R.; Beckenhauer, H.C.; Sorrell, M.F.; Tuma, D.J. Betaine attenuates alcoholic steatosis by restoring phosphatidylcholine generation via the phosphatidylethanolamine methyltransferase pathway. J. Hepatol. 2007, 46, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Lieber, C.S.; Robins, S.J.; Li, J.; DeCarli, L.M.; Mak, K.M.; Fasulo, J.M.; Leo, M.A. Phosphatidylcholine protects against fibrosis and cirrhosis in the baboon. Gastroenterology 1994, 106, 152–159. [Google Scholar] [CrossRef]
- Ooi, K.; Shiraki, K.; Sakurai, Y.; Morishita, Y.; Nobori, T. Clinical significance of abnormal lipoprotein patterns in liver diseases. Int. J. Mol. Med. 2005, 15, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Chrostek, L.; Supronowicz, L.; Panasiuk, A.; Cylwik, B.; Gruszewska, E.; Flisiak, R. The effect of the severity of liver cirrhosis on the level of lipids and lipoproteins. Clin. Exp. Med. 2014, 14, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Nilsson-Ehle, P.; Xu, N. Influence of liver cancer on lipid and lipoprotein metabolism. Lipids Health Dis. 2006, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Bassani, L.; Fernandes, S.A.; Raimundo, F.V.; Harter, D.L.; Gonzalez, M.C.; Marroni, C.A. Lipid Profile of Cirrhotic Patients and Its Association with Prognostic Scores: A cross-sectional study. Arq. Gastroenterol. 2015, 52, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Obeid, R.; Herrmann, W. Homocysteine and lipids: S-adenosyl methionine as a key intermediate. FEBS Lett. 2009, 583, 1215–1225. [Google Scholar] [CrossRef] [PubMed]
- Namekata, K.; Enokido, Y.; Ishii, I.; Nagai, Y.; Harada, T.; Kimura, H. Abnormal lipid metabolism in cystathionine beta-synthase-deficient mice, an animal model for hyperhomocysteinemia. J. Biol. Chem. 2004, 279, 52961–52969. [Google Scholar] [CrossRef] [PubMed]
- Mikael, L.G.; Genest, J., Jr.; Rozen, R. Elevated homocysteine reduces apolipoprotein A-I expression in hyperhomocysteinemic mice and in males with coronary artery disease. Circ. Res. 2006, 98, 564–571. [Google Scholar] [CrossRef] [PubMed]
- McKay, J.A.; Xie, L.; Adriaens, M.; Evelo, C.T.; Ford, D.; Mathers, J.C. Maternal folate depletion during early development and high fat feeding from weaning elicit similar changes in gene expression, but not in DNA methylation, in adult offspring. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Wankhade, U.D.; Zhong, Y.; Kang, P.; Alfaro, M.; Chintapalli, S.V.; Thakali, K.M.; Shankar, K. Enhanced offspring predisposition to steatohepatitis with maternal high-fat diet is associated with epigenetic and microbiome alterations. PLoS ONE 2017, 12, e0175675. [Google Scholar] [CrossRef] [PubMed]
- Souza-Mello, V.; Mandarim-de-Lacerda, C.A.; Aguila, M.B. Hepatic structural alteration in adult programmed offspring (severe maternal protein restriction) is aggravated by post-weaning high-fat diet. Br. J. Nutr. 2007, 98, 1159–1169. [Google Scholar] [CrossRef] [PubMed]

| Diet (Maternal/Offspring) | Two-Way ANOVA | ||||
|---|---|---|---|---|---|
| CAS/CON | CAS/EtOH | SPI/CON | SPI/EtOH | ||
| Body weight (g) | |||||
| 3 weeks | 62.1 ± 1.0 a | 64.6 ± 1.5 a | 54.3 ± 2.1 b | 55.1 ± 1.8 b | M |
| 8 weeks | 400.1 ± 6.1 | 400.1 ± 4.8 | 385.6 ± 10.1 | 383.1 ± 10.2 | |
| 15 weeks(at sacrifice) | 533.4 ± 12.3 | 497.9 ± 10.8 | 531.8 ± 22.8 | 511.6 ± 20.0 | |
| Organ weight (g) | |||||
| Liver | 15.0 ± 0.4 | 16.9 ± 0.8 | 15.3 ± 1.3 | 17.0 ± 1.3 | |
| Epididymal fat | 14.0 ± 1.3 | 10.7 ± 0.7 | 12.7 ± 1.1 | 10.4 ± 1.6 | E |
| Relative liver weight (g/100 g body weight) | |||||
| Liver | 2.8 ± 0.1 b | 3.4 ± 0.1 a | 2.9 ± 0.1 b | 3.3 ± 0.1 a | E |
| Epididymal fat | 2.6 ± 0.2 a | 2.1 ± 0.1 a,b | 2.4 ± 0.1 a,b | 2.0 ± 0.2 b | E |
| Hepatic Gene Expression (mRNA) | Correlation Coefficient (r) | p-Value |
|---|---|---|
| GNMT | −0.341 | 0.141 |
| PEMT | −0.457 | 0.043 |
| BHMT | −0.599 | 0.005 |
| MTR | −0.690 | 0.001 |
| CBS | −0.631 | 0.003 |
| Hepatic Gene Expression (mRNA) | Correlation Coefficient (r) | p-Value |
|---|---|---|
| ABCA1 | –0.594 | 0.006 |
| ABCG5 | –0.544 | 0.016 |
| LCAT | –0.754 | < 0.001 |
| LDLR | –0.697 | 0.001 |
| LXRα | –0.631 | 0.003 |
| SRB1 | –0.542 | 0.020 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Won, S.B.; Kwon, Y.H. Maternal Consumption of Low-Isoflavone Soy Protein Isolate Confers the Increased Predisposition to Alcoholic Liver Injury in Adult Rat Offspring. Nutrients 2018, 10, 332. https://doi.org/10.3390/nu10030332
Won SB, Kwon YH. Maternal Consumption of Low-Isoflavone Soy Protein Isolate Confers the Increased Predisposition to Alcoholic Liver Injury in Adult Rat Offspring. Nutrients. 2018; 10(3):332. https://doi.org/10.3390/nu10030332
Chicago/Turabian StyleWon, Sae Bom, and Young Hye Kwon. 2018. "Maternal Consumption of Low-Isoflavone Soy Protein Isolate Confers the Increased Predisposition to Alcoholic Liver Injury in Adult Rat Offspring" Nutrients 10, no. 3: 332. https://doi.org/10.3390/nu10030332




