A Comprehensive Peptidomic Approach to Characterize the Protein Profile of Selected Durum Wheat Genotypes: Implication for Coeliac Disease and Wheat Allergy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
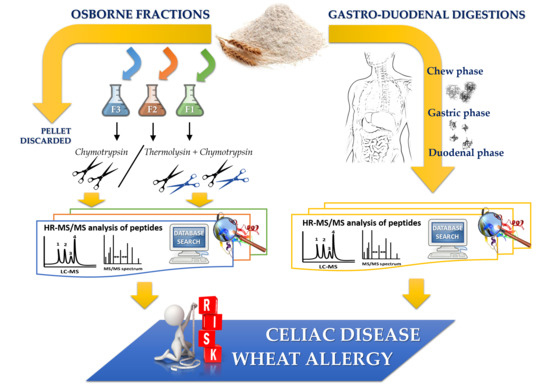
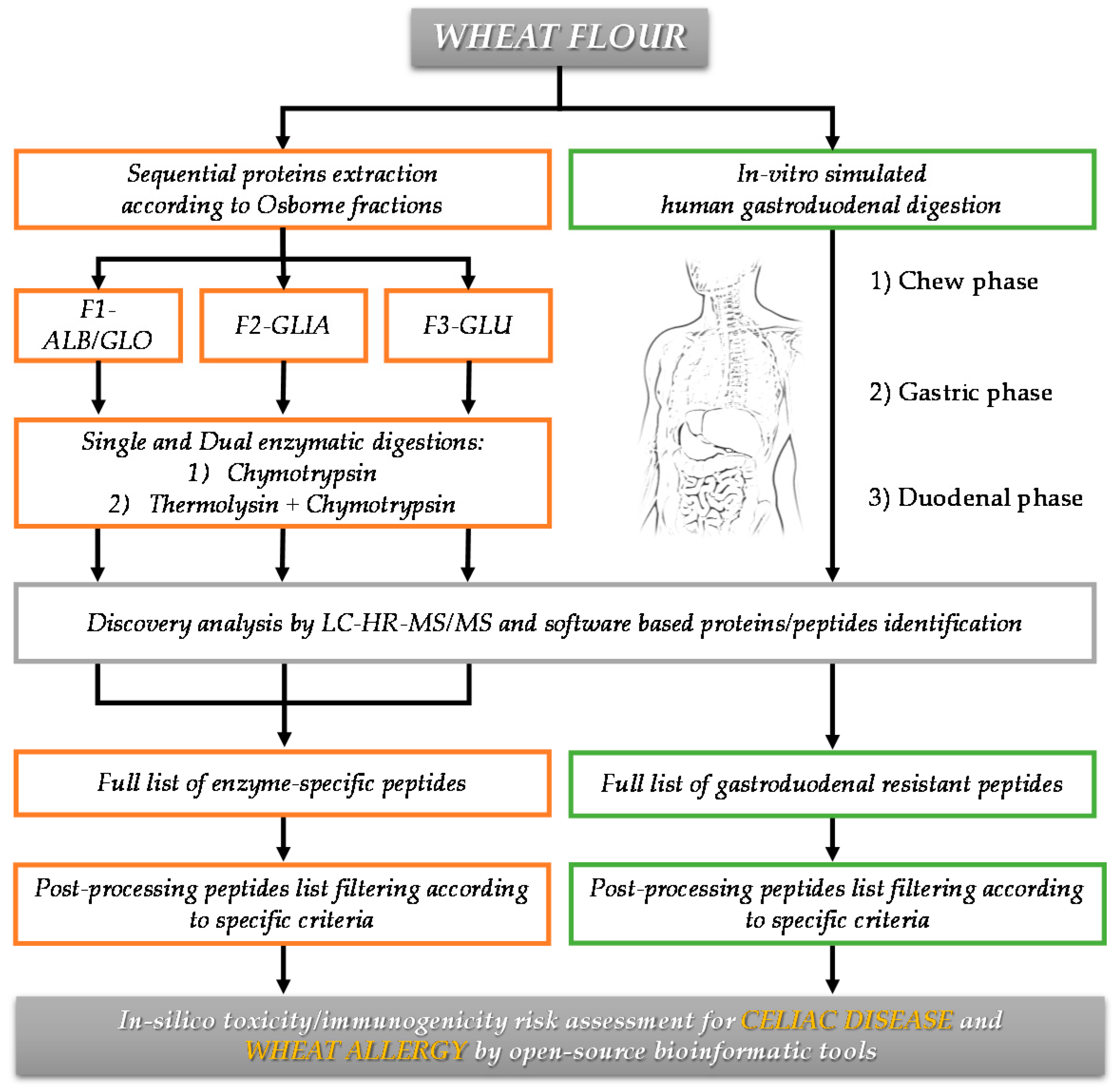
2.2. Sequential Protein Extraction of Osborne Fractions and Total Protein Quantification
2.3. Bottom-Up Approach for Protein Characterization
2.3.1. Single Enzyme Digestion
2.3.2. Dual Enzyme Digestion
2.4. SDS-PAGE Analysis
2.5. In Vitro-Simulated Human Gastroduodenal Digestion Experiments
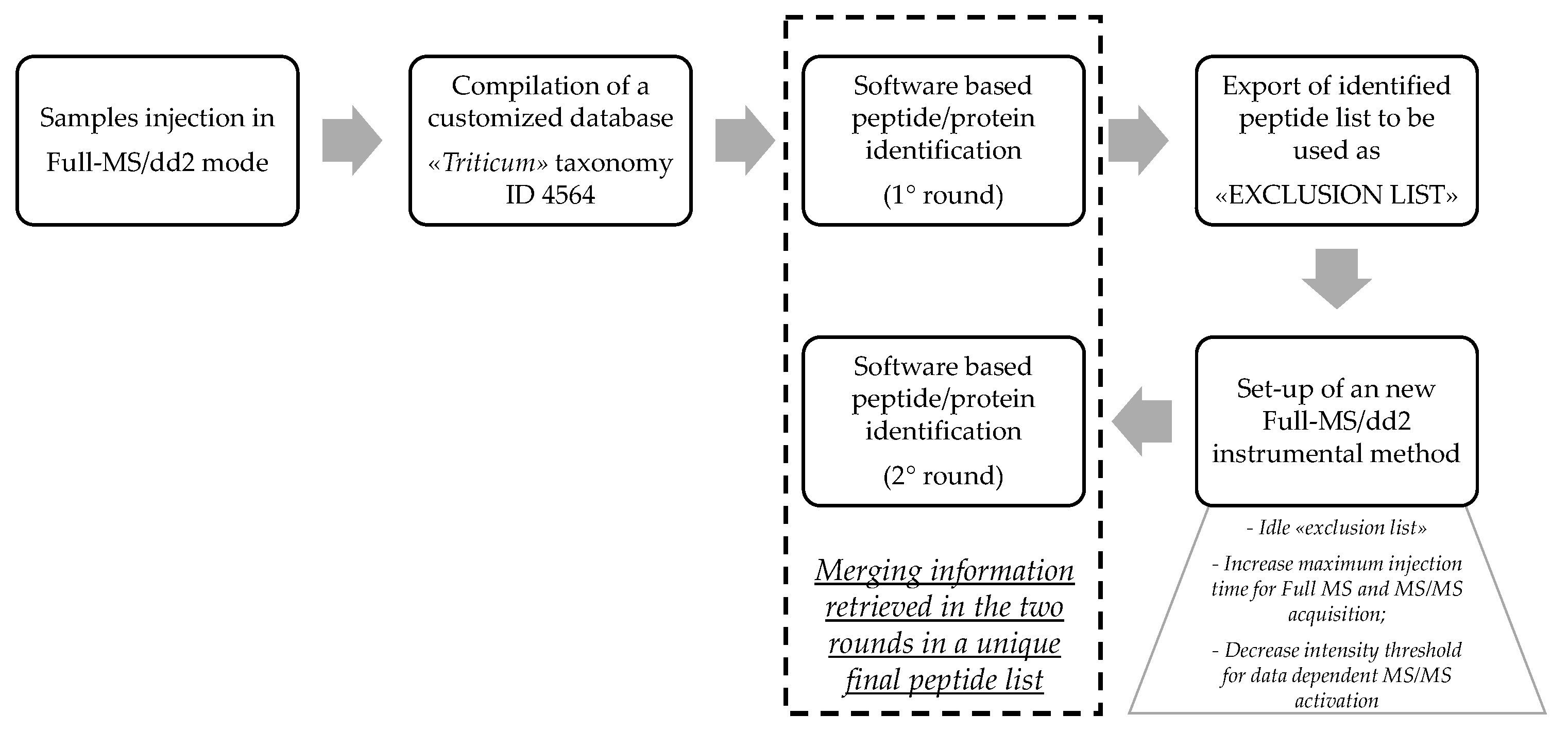
2.6. Discovery HR-MS/MS Analysis and Peptide Identification
2.7. In Silico Toxicity/Immunogenicity Risk Assessment
3. Results and Discussion
3.1. Specific Enzymatic Digestion of Osborne Protein Fractions
3.2. In Vitro-Simulated Human Gastroduodenal Digestion Experiments
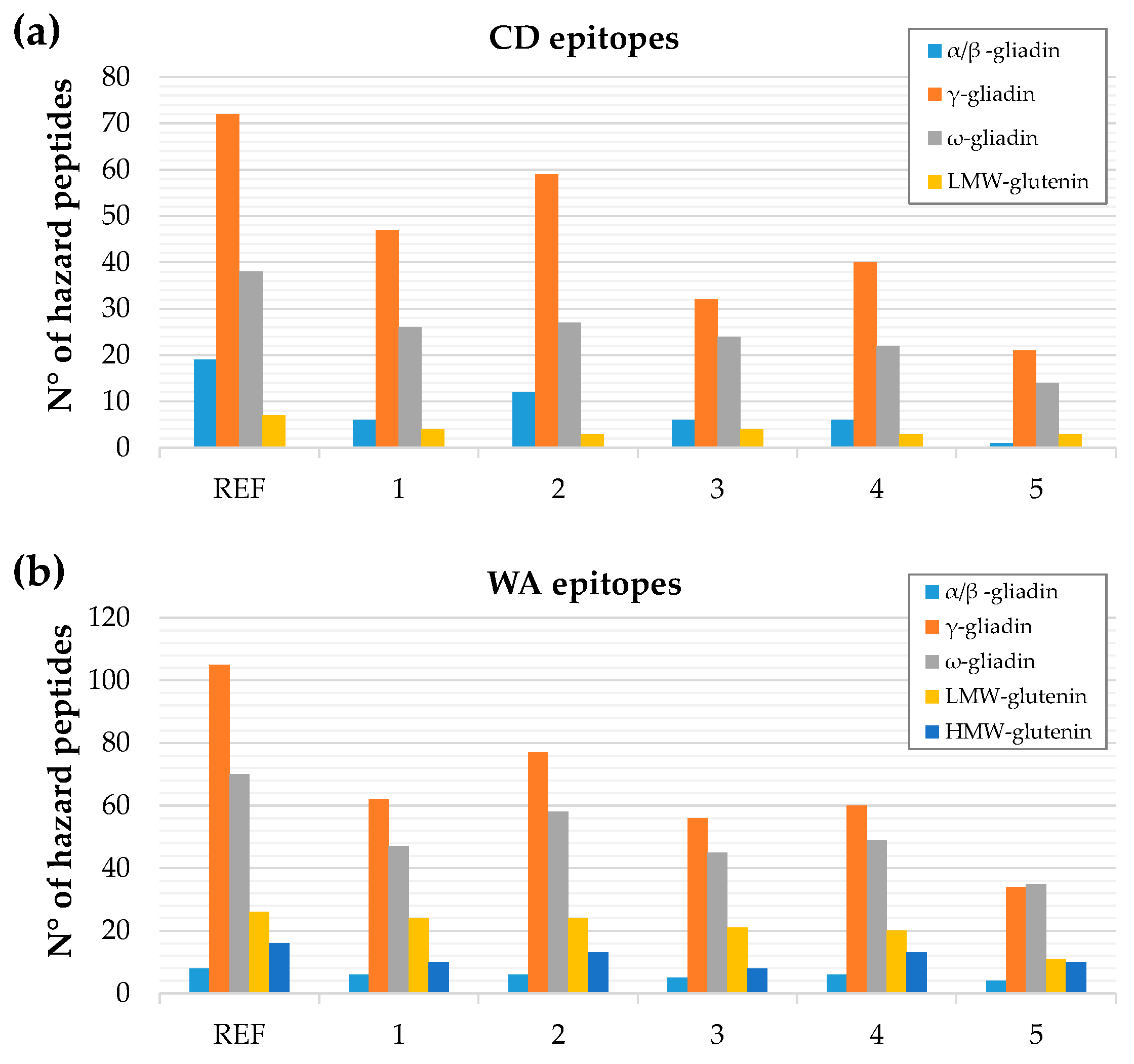
3.3. In Silico Toxicity/Immunogenicity Risk Assessment and Implication for Celiac Disease and Wheat Allergy Patients
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Alves, T.O.; D’Almeida, C.T.S.; Victorio, V.C.M.; Souz, G.H.M.F.; Cameron, L.C.; Ferreira, M.S.L. Immunogenic and allergenic profile of wheat flours from different technological qualities revealed by ion mobility mass spectrometry. J. Food Compos. Anal. 2018, 73, 67–75. [Google Scholar] [CrossRef]
- Goesaert, H.; Brijs, K.; Veraverbeke, W.S.; Courtin, C.M.; Gebruers, K.; Delcour, J.A. Wheat flour constituents: How they impact bread quality, and how to impact their functionality. Trends Food Sci. Technol. 2005, 16, 12–30. [Google Scholar] [CrossRef]
- Gao, L.; Wang, A.; Li, X.; Dong, K.; Wang, K.; Appels, R.; Ma, W.; Yan, Y. Wheat quality related differential expressions of albumins and globulins revealed by twodimensional difference gel electrophoresis (2-D DIGE). J. Proteom. 2009, 73, 279–296. [Google Scholar] [CrossRef] [PubMed]
- D’Ovidio, R.; Masci, S. The low-molecular-weight glutenin subunits of wheat gluten. J. Cereal Sci. 2004, 39, 321–339. [Google Scholar] [CrossRef]
- Uvackova, L.; Skultety, L.; Bekesova, S.; McClain, S.; Hajduch, M. MSE based multiplex protein analysis quantified important allergenic proteins and detected relevant peptides carrying known epitopes in wheat grain extracts. J. Proteome Res. 2013, 12, 4862–4869. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R.; Tatham, A.S. Improving wheat to remove coeliac epitopes but retain functionality. J. Cereal Sci. 2016, 67, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Sollid, L.M.; Qiao, S.-W.; Anderson, R.P.; Gianfrani, C.; Konig, F. Nomenclature and listings of celiac disease relevant gluten T-cell epotopies restricted by HLA-DQ molecules. Immunogenetics 2012, 64, 455–460. [Google Scholar] [CrossRef]
- Bouchez-Mahiout, I.; Snégaroff, J.; Tylichova, M.; Pecquet, C.; Branlard, G.; Laurière, M. Low molecular weight glutenins in wheat-dependant, exercise-induced anaphylaxis: Allergenicity and antigenic relationships with omega 5-gliadins. Int. Arch. Allergy Immunol. 2010, 153, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, S.C.; Fischer, J.; Eriksson, C.; Bengtsson Gref, O.; Biedermann, T.; Jakob, T. IgE detection to α/β/γ-gliadin and its clinical relevance in wheat-dependent exercise-induced anaphylaxis. Allergy 2012, 67, 1457–1460. [Google Scholar] [CrossRef]
- Matsuo, H.; Kohno, K.; Niihara, H.; Morita, E. Specific IgE determination to epitope peptides of ω-5 gliadin and high molecular weight glutenin subunit is a useful tool for diagnosis of wheat-dependent exercise-induced anaphylaxis. J. Immunol. 2005, 175, 8116–8122. [Google Scholar] [CrossRef]
- Burkhardt, J.G.; Chapa-Rodriguez, A.; Bahna, S.L. Gluten sensitivities and the allergist: Threshing the grain from the husks. Allergy 2018, 73, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Mamone, G.; Picariello, G.; Addeo, F.; Ferranti, P. Proteomic analysis in allergy and intolerance to wheat products. Expert Rev. Proteom. 2011, 8, 95–115. [Google Scholar] [CrossRef] [PubMed]
- Boukid, F.; Mejri, M.; Pellegrini, N.; Sforza, S.; Prandi, B. How Looking for celiac-safe wheat can influence its technological properties. Compr. Rev. Food Sci. Food Saf. 2017, 16, 797–807. [Google Scholar] [CrossRef]
- Pilolli, R.; Gadaleta, A.; Mamone, G.; Nigro, D.; De Angelis, E.; Montemurro, N.; Monaci, L. Scouting for naturally low-toxicity wheat genotypes by a multidisciplinary approach. Sci. Rep. 2019, 9, 1646. [Google Scholar] [CrossRef] [PubMed]
- Laidò, G.; Mangini, G.; Taranto, F.; Gadaleta, A.; Blanco, A.; Cattivelli, L.; Marone, D.; Mastrangelo, A.M.; Papa, R.; De Vita, P. Genetic Diversity and Population Structure of Tetraploid Wheats (Triticum turgidum L.) Estimated by SSR, DArT and Pedigree Data. PLoS ONE 2013, 8, e67280. [Google Scholar] [CrossRef] [PubMed]
- Minekus, M.; Alminger, M.; Alvito, P.; Balance, S.; Bohn, T.; Bourlieu, C.; Carriere, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised static in vitro digestion method suitable for food an international consensus. Food Funct. 2014, 56, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Weiss, H.; Antes, S.; Seilmeier, W. Quantitative determination of gluten protein types in wheat flour by reversed-phase high performance liquid chromatography. Cereal Chem. 1998, 75, 644–650. [Google Scholar] [CrossRef]
- De Angelis, E.; Bavaro, S.L.; Forte, G.; Pilolli, R.; Monaci, L. Heat and Pressure Treatments on Almond Protein Stability and Change in Immunoreactivity after Simulated Human Digestion. Nutrients 2018, 10, 1679. [Google Scholar] [CrossRef]
- De Angelis, E.; Pilolli, R.; Bavaro, S.L.; Monaci, L. Insight into the gastro-duodenal digestion resistance of soybean proteins and potential implications for residual immunogenicity. Food Funct 2017, 8, 1599–1610. [Google Scholar] [CrossRef]
- Uniprot Database. Available online: https://www.uniprot.org (accessed on 23 March 2019).
- Celiac Disease (CD) Novel Protein Risk Assessment Tool. Available online: http://www.allergenonline.org/celiachome.shtml (accessed on 10 June 2019).
- Immune Epitope Database and Analysis Resource. Available online: https://www.iedb.org/ (accessed on 30 May 2019).
- Jouanin, A.; Gilissen, L.J.W.J.; Boyd, L.A.; Cockram, J.; Leigh, F.J.; Wallington, E.J.; van den Broeck, H.C.; van der Meer, I.M.; Schaart, J.G.; Visser, R.G.F.; et al. Food processing and breeding strategies for coeliac-safe and healthy wheat products. Food Res. Int. 2018, 110, 11–21. [Google Scholar] [CrossRef] [Green Version]
- Shewry, P.R.; D’Ovidio, R.; Lafiandra, D.; Jenkins, J.A.; Mills, E.N.C.; Békés, F. Wheat Grain Proteins. In Wheat Chemistry and Technology, 4th ed.; Khan, K., Shewry, P.R., Eds.; American Association of Cereal Chemists International Press: Saint Paul, MN, USA, 2009; pp. 223–298. [Google Scholar]
- Mamone, G.; Nitride, C.; Picariello, G.; Addeo, F.; Ferranti, P.; Mackie, A. Tracking the fate of pasta (T. Durum semolina) immunogenic proteins by in vitro simulated digestion. J. Agric. Food Chem. 2015, 63, 2660–2667. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Genetically Modified Organisms (GMO). Guidance on allergenicity assessment of genetically modified plants. EFSA J. 2017, 15, e04862. [Google Scholar] [CrossRef]
- Mohan, J.F.; Unanue, E.R. Unconventional recognition of peptides by T cells and the implications for autoimmunity. Nat. Rev. Immunol. 2012, 12, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Gianfrani, C.; Camarca, A.; Mazzarella, G.; Di Stasio, L.; Giardullo, N.; Ferranti, P.; Picariello, G.; Rotondi Aufiero, V.; Picascia, S.; Troncone, R.; et al. Extensive in vitro gastrointestinal digestion markedly reduces the immune-toxicity of Triticum monococcum wheat: Implication for celiac disease. Mol. Nutr. Food Res. 2015, 59, 1844–1854. [Google Scholar] [CrossRef] [PubMed]
- Pisapia, L.; Camarca, A.; Picascia, S.; Bassi, V.; Barba, P.; Del Pozzo, G.; Gianfrani, C. HLA-DQ2.5 genes associated with celiac disease risk are preferentially expressed with respect to non-predisposing HLA genes: Implication for anti-gluten T cell response. J. Autoimmun. 2016, 70, 63–72. [Google Scholar] [CrossRef]
- Gianfrani, C.; Pisapia, L.; Picascia, S.; Strazzullo, M.; Del Pozzo, G. Expression level of risk genes of MHC class II is a susceptibility factor for autoimmunity: New insights. J. Autoimmun. 2018, 89, 1–10. [Google Scholar] [CrossRef] [PubMed]


| Sample Code | Accession | Taxonomic Classification | Year of Release | C/NC | Origin |
|---|---|---|---|---|---|
| 1 | Duetto | T. turgidum ssp. durum | 2002 | C | Italy |
| 2 | Colosseo | T. turgidum ssp. durum | 1995 | C | Italy |
| 3 | Lloyd | T. turgidum ssp. durum | 1983 | C | United States |
| 4 | Neolatino | T. turgidum ssp. durum | 2007 | C | Italy |
| 5 | PI 56263 | T. turgidum ssp. turgidum | - | NC | Portugal, Lisboa |
| Description | Fractions | Features | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|---|---|
| Single enzyme digests (CHY) | F1+F2+F3 | # Proteins | 359 | 373 | 339 | 395 | 276 |
| # Peptides | 672 | 693 | 680 | 747 | 590 | ||
| Multiconsensus of Single + Dual enzyme digests (CHY + THE/CHY) | F1+F2+F3 | # Proteins | 420 | 411 | 413 | 438 | 361 |
| # Peptides | 719 | 777 | 767 | 814 | 698 |
| Description | REF | 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|---|---|
| Total No. of identified peptides 1 (sequence length cut-off 9AA) | Abs | 2027 | 1578 | 1732 | 1554 | 1513 | 1365 |
| No. peptides containing intact epitopes relevant for CD | Abs | 118 | 69 | 87 | 53 | 60 | 32 |
| Rel | 100% | 58% | 74% | 45% | 51% | 27% | |
| Total No. of identified peptides 1 (sequence length cut-off 5AA) | Abs | 2955 | 2469 | 2629 | 2440 | 2354 | 2136 |
| No. peptides containing intact epitopes relevant for WA | Abs | 208 | 133 | 165 | 121 | 136 | 86 |
| Rel | 100% | 64% | 79% | 58% | 65% | 41% | |
| CD Epitope Sequence (ID Number) * | HLA-DQ Molecules * | Protein * | 9AA Restricted Core Epitope | Sample | |||||
|---|---|---|---|---|---|---|---|---|---|
| REF | 1 | 2 | 3 | 4 | 5 | ||||
| PQPQPFPSQQPY (7) | HLA-DR | α-gliadin | - | X | X | X | X | X | |
| QPFPQPQLPY (42) | DQ2 | α-gliadin | DQ2.5-glia-α1a | X | X | X | X | ||
| PFPQPQLPYPQ (45) | DQ2 | α-gliadin | DQ2.5-glia-α1a DQ2.5-glia-α2 | X | X | ||||
| PFPQPQLPY (53) | DQ2.5 | α-9 gliadin | DQ2.5-glia-α1a | X | X | X | X | ||
| PQPQLPYPQPQL (64) | DQ2 | α-gliadin | DQ2.5-glia-α2 | X | X | ||||
| FRPQQPYPQ (93) | DQ2.5 | α-20 gliadin | DQ2.5-glia-α3 | X | X | X | X | X | |
| PQQPYPQPQPQ (138) | DQ2 | α-gliadin | - | X | |||||
| LGQQQPFPPQQPYPQPQ (151) | DQ2 (α1 * 0501, α1 * 0201) | α-gliadin | - | X | X | X | X | ||
| LGQQQPFPPQQPY (152) | HLA-DR | α-gliadin | - | X | X | X | X | ||
| QPFPQPQLPYSQ (164) | DQ2 | α-gliadin | DQ2.5-glia-α1a | X | X | X | X | ||
| PFPQPQLPYSQ (166) | DQ2 | α-gliadin | DQ2.5-glia-α1a | X | X | X | X | ||
| QPQPFLPQLPYPQP (185) | DQ2 | α-gliadin | - | X | X | ||||
| PQPFLPQLPYPQ (187) | DQ2 | α-gliadin | - | X | X | X | X | X | |
| PLQPQQPFPQQPQQPFPQPQ (224) | DQ2 | ω-gliadin | DQ2.5-glia-γ4c DQ2.5-glia-γ5 DQ8-glia-γ1a | X | |||||
| PQQPQQPFPLQPQQPFPQQP (235) | DQ8 | ω-gliadin | - | X | |||||
| QPFPLQPQQPVPQQPQ (976) | DQ2 | ω-gliadin | - | X | X | X | X | ||
| PFPQPQQPF (867) | DQ2.5 | hor-1 | DQ2.5-glia-ω1 DQ2.5-hor-1 DQ2.5-sec-1 | X | X | X | X | X | |
| PQPQQPFPQ (891) | DQ2.5 | hor-2 | DQ2.5-hor-2 DQ2.5-sec-2 | X | X | X | X | X | |
| PQQPFPQPQQPFPQ (911) | DQ2 | ω-gliadin | DQ2.5-glia-ω1 DQ2.5-hor-1 DQ2.5-hor-2 DQ2.5-sec-1 DQ2.5-sec-2 | X | X | X | X | X | |
| PQTQQPQQPFPQ (926) | DQ2 | γ-gliadin | DQ2.5-glia-γ4c DQ8-glia-γ1a | X | X | X | X | X | |
| QSIPQPQQPFPQ (930) | DQ2 | γ-gliadin | DQ2.5-hor-2 DQ2.5-sec-2 | X | X | X | X | ||
| QPFPQPQQPFPQ (935) | DQ2 | γ-gliadin | DQ2.5-glia-ω1 DQ2.5-hor-1 DQ2.5-hor-2 DQ2.5-sec-1 DQ2.5-sec-2 | X | X | X | X | X | |
| QQPQQPYPQ (458) | DQ2.5/DQ8 | γ-1 and γ-5 gliadin | DQ2.5-glia-γ3 DQ8-glia-γ1b | X | X | X | |||
| QQPYPQQPQ (464) | DQ2 | γ-gliadin | - | X | X | ||||
| PYPQQPQQP (468) | DQ2 | γ-gliadin | - | X | X | ||||
| PFPQPQQTFPQQPQLPFPQQ (502) | DQ2, DQ8 | γ1-gliadin | - | X | X | X | X | ||
| PFPQPQQTFPQ (503) | DQ2 | γ1-gliadin | - | X | X | X | X | ||
| PQQTFPQQPQLP (504) | DQ2 | γ1-gliadin | - | X | X | X | X | ||
| TQQPQQPFPQP (534) | DQ2 | γ-gliadin | DQ2.5-glia-γ4c DQ8-glia-γ1a | X | X | X | X | ||
| PFPQTQQPQQPFPQ (553) | DQ8 (DQ2/8) | γ-gliadin | DQ2.5-glia-γ4c DQ8-glia-γ1a | X | X | X | X | X | |
| PFPQPQQPQQPFPQ (644) | DQ8 (DQ2/8) | γ-gliadin | DQ2.5-glia-γ4c DQ8-glia-γ1a | X | |||||
| QPFPQLQQPQQP (650) | DQ2 | γ-gliadin | - | X | |||||
| QQPPFSQQQQPVLPQ (701) | DQ2 | γ-gliadin or LMW glutenin | DQ2.2-glut-L1 | X | X | X | X | X | |
| QQPPFSQQQQPQFSQ (751) | DQ2 | LMW glutenin | - | X | X | X | X | X | |
| Protein (Allergen Code) | WA Epitope Sequence (ID Number *) | Samples | |||||
|---|---|---|---|---|---|---|---|
| REF | 1 | 2 | 3 | 4 | 5 | ||
| α-gliadin (Tri a 21) | YLQLQPFPQP (148993) | X | X | X | X | X | |
| γ-gliadin (Tri a 20) | PQQPFPQLQQ (148736) | X | X | X | X | X | |
| γ-gliadin (Tri a 20) | QQQLPQPQQP (148860) | X | X | X | X | ||
| γ-gliadin (Tri a 20) | QQPVPQPHQPFSQQ (192607) | X | |||||
| ω-gliadin (Tri a 19) | QQPQQPFPLQ (52105) | X | X | X | X | X | |
| ω-gliadin (Tri a 19) | FPQQQFPQQQ (148610) | X | X | ||||
| ω-gliadin (Tri a 19) | PFPQPQQPFP (148713) | X | X | X | X | X | |
| ω-gliadin (Tri a 19) | PQQSPEQQQF (148748) | X | |||||
| ω-gliadin (Tri a 19) | QFPQQQFPQQ (148762) | X | X | ||||
| ω-gliadin (Tri a 19) | QQFPQQQFPQ (148828) | X | X | X | |||
| ω-gliadin (Tri a 19) | QQLQQPFPLQ (148834) | X | X | X | X | ||
| ω-gliadin (Tri a 19) | QQPIPVQPQQ (148841) | X | X | X | X | X | |
| ω-gliadin (Tri a 19) | QQPQQPFPQL (148843) | X | |||||
| ω-gliadin (Tri a 19) | QQQFPQQPPQ (148856) | X | X | ||||
| ω-gliadin (Tri a 19) | QQQFPQQQFP (148857) | X | X | X | |||
| ω-gliadin (Tri a 19) | QSPEQQQFPQ (148898) | X | |||||
| ω-gliadin (Tri a 19) | FPQQQFPQQQFPQ (173928) | X | X | ||||
| ω-gliadin (Tri a 19) | PQQQFPQQQQFPQ (173960) | X | |||||
| ω-gliadin (Tri a 19) | PQQSPEQQQFPQQ (173967) | X | |||||
| ω-gliadin (Tri a 19) | QQQFPQQQFPQQP (173992) | X | X | ||||
| ω-gliadin (Tri a 19) | EQQQFPQQQF (190740) | X | |||||
| ω-gliadin (Tri a 19) | PQQPQQFPQQ (190898) | X | X | X | |||
| ω-gliadin (Tri a 19) | PQQQQFPQQQ (190900) | X | X | X | X | X | |
| ω-gliadin (Tri a 19) | QPQQFPQQQF (190924) | X | X | ||||
| ω-gliadin (Tri a 19) | QQFPQQQQFP (190931) | X | X | X | X | X | |
| ω-gliadin (Tri a 19) | QQFPQQQQLP (190932) | X | X | ||||
| ω-gliadin (Tri a 19)/γ-gliadin (Tri a 20) | QTQQPQQPFP (52600) | X | X | X | X | X | |
| HMW-glutenin (Tri a 26) | QPGQGQQGQQPGQG (113765) | X | X | X | X | X | |
| HMW-glutenin (Tri a 26) | QPGQGQQPGQGQPG (113766) | X | |||||
| HMW-glutenin (Tri a 26) | QQPGQGQQPGQGQQ (113781) | X | |||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pilolli, R.; Gadaleta, A.; Di Stasio, L.; Lamonaca, A.; De Angelis, E.; Nigro, D.; De Angelis, M.; Mamone, G.; Monaci, L. A Comprehensive Peptidomic Approach to Characterize the Protein Profile of Selected Durum Wheat Genotypes: Implication for Coeliac Disease and Wheat Allergy. Nutrients 2019, 11, 2321. https://doi.org/10.3390/nu11102321
Pilolli R, Gadaleta A, Di Stasio L, Lamonaca A, De Angelis E, Nigro D, De Angelis M, Mamone G, Monaci L. A Comprehensive Peptidomic Approach to Characterize the Protein Profile of Selected Durum Wheat Genotypes: Implication for Coeliac Disease and Wheat Allergy. Nutrients. 2019; 11(10):2321. https://doi.org/10.3390/nu11102321
Chicago/Turabian StylePilolli, Rosa, Agata Gadaleta, Luigia Di Stasio, Antonella Lamonaca, Elisabetta De Angelis, Domenica Nigro, Maria De Angelis, Gianfranco Mamone, and Linda Monaci. 2019. "A Comprehensive Peptidomic Approach to Characterize the Protein Profile of Selected Durum Wheat Genotypes: Implication for Coeliac Disease and Wheat Allergy" Nutrients 11, no. 10: 2321. https://doi.org/10.3390/nu11102321
APA StylePilolli, R., Gadaleta, A., Di Stasio, L., Lamonaca, A., De Angelis, E., Nigro, D., De Angelis, M., Mamone, G., & Monaci, L. (2019). A Comprehensive Peptidomic Approach to Characterize the Protein Profile of Selected Durum Wheat Genotypes: Implication for Coeliac Disease and Wheat Allergy. Nutrients, 11(10), 2321. https://doi.org/10.3390/nu11102321