No Influence of the Fat Mass and Obesity-Associated Gene rs9939609 Single Nucleotide Polymorphism on Blood Lipids in Young Males
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Preliminary Measures
2.3. Experimental Procedure
2.4. Blood Sampling
2.5. Biochemical Analysis
2.6. Statistical Analyses
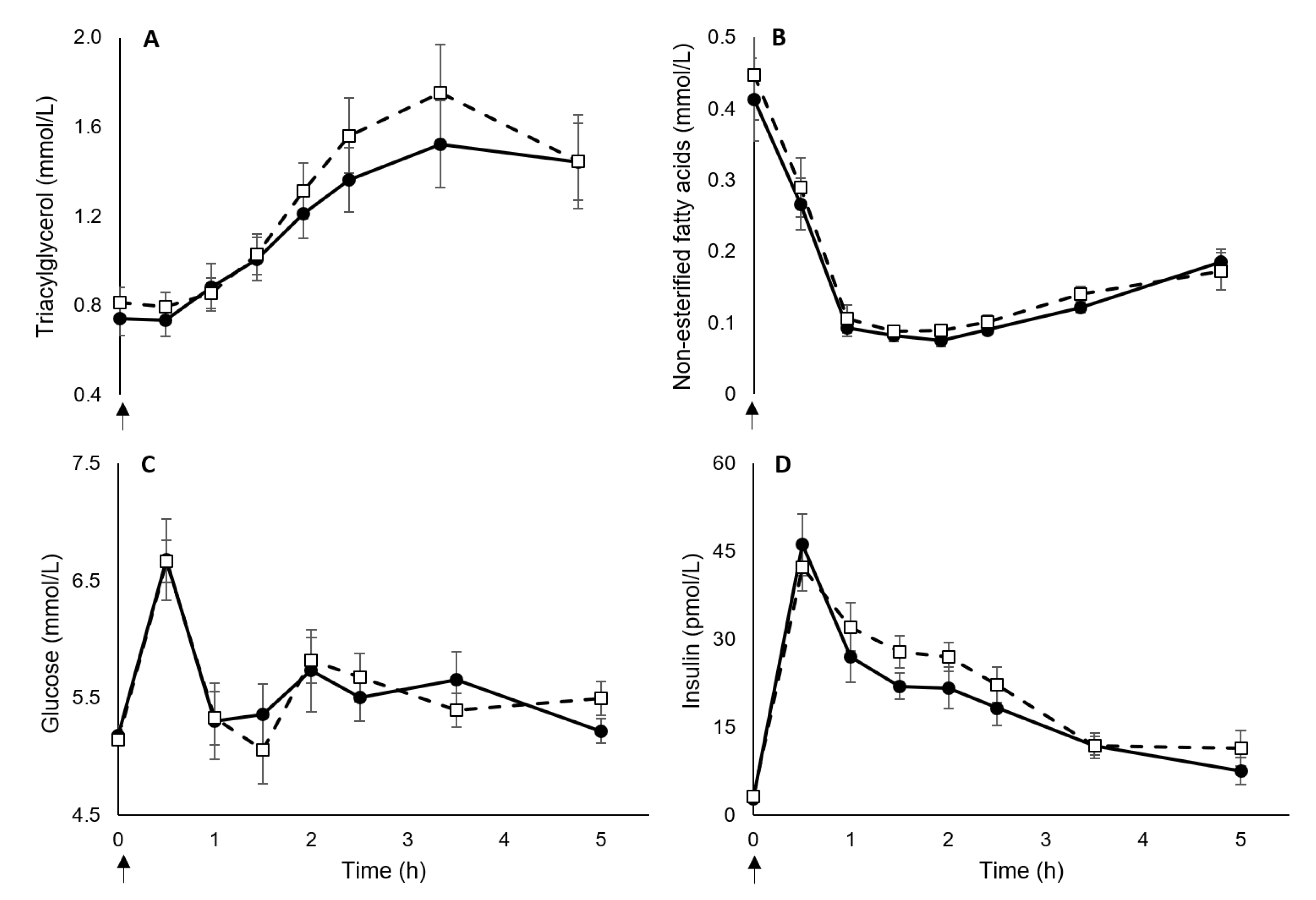
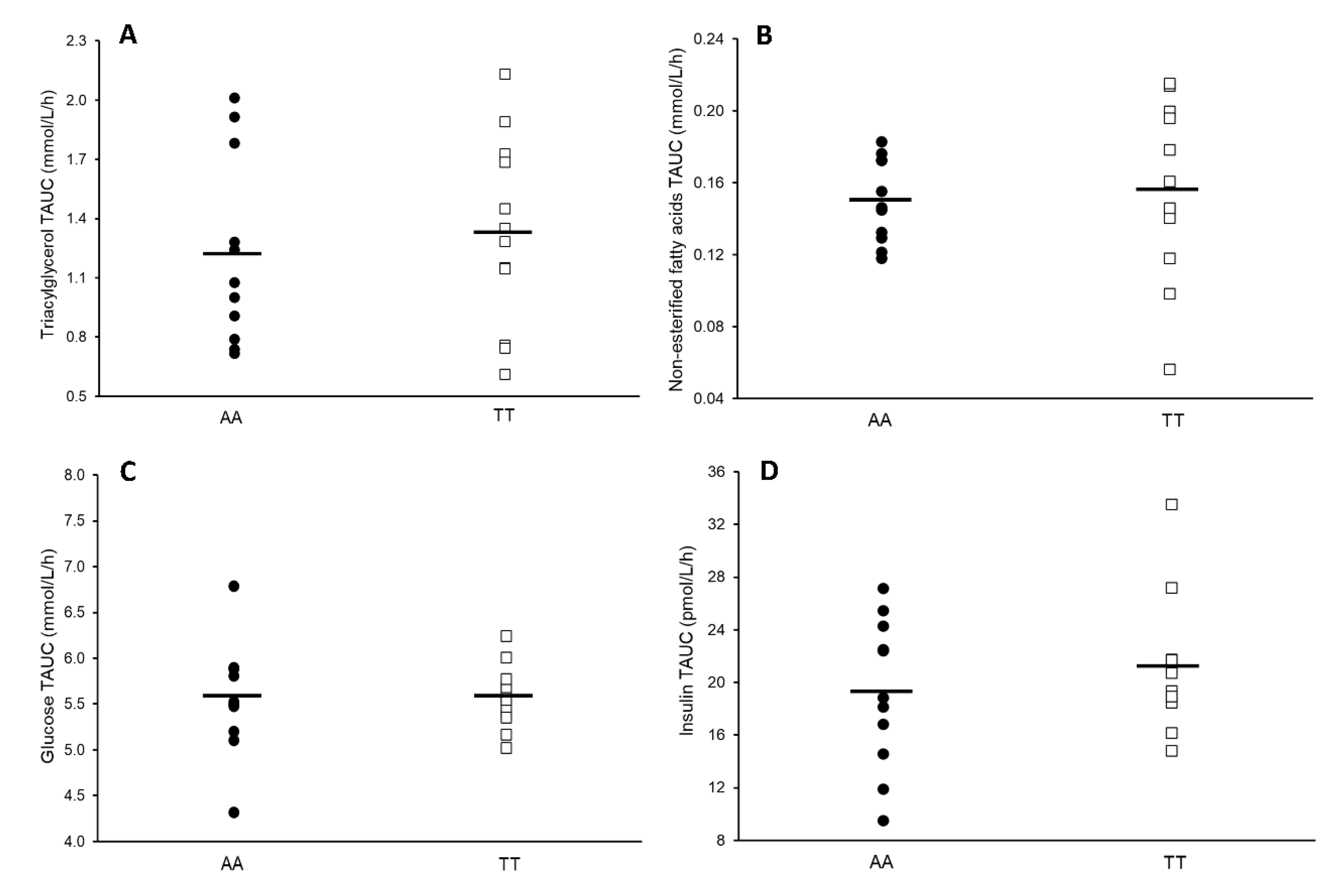
3. Results
3.1. Participant Characteristics
3.2. Blood Markers
3.3. Retrospective Power Calculations
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.B.; Elliott, K.S.; Lango, H.; Rayner, N.W.; et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loos, R.J.F.; Yeo, G.S.H. The bigger picture of FTO: The first GWAS-identified obesity gene. Nat. Rev. Endocrinol. 2014, 10, 51–61. [Google Scholar] [CrossRef]
- Wardle, J.; Llewellyn, C.; Sanderson, S.; Plomin, R. The FTO gene and measured food intake in children. Int. J. Obes. 2009, 33, 42–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karra, E.; Daly, O.G.O.; Choudhury, A.I.; Yousseif, A.; Millership, S.; Neary, M.T.; Scott, W.R.; Chandarana, K.; Manning, S.; Hess, M.E.; et al. A link between FTO, ghrelin, and impaired brain food-cue responsivity. J. Clin. Investig. 2013, 123, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Mou, S.; Pan, C. The FTO gene rs9939609 polymorphism predicts risk of cardiovascular disease: A systematic review and meta-analysis. PLoS ONE 2013, 8, e71901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Äijälä, M.; Ronkainen, J.; Huusko, T.; Malo, E.; Savolainen, E.R.; Savolainen, M.J.; Salonurmi, T.; Bloigu, R.; Kesäniemi, Y.A.; Ukkola, O. The fat mass and obesity-associated (FTO) gene variant rs9939609 predicts long-term incidence of cardiovascular disease and related death independent of the traditional risk factors. Ann. Med. 2015, 47, 655–663. [Google Scholar] [CrossRef]
- Doney, A.S.F.; Dannfald, J.; Kimber, C.H.; Donnelly, L.A.; Pearson, E.; Morris, A.D.; Palmer, C.N.A. The FTO gene is associated with an atherogenic lipid profile and myocardial infarction in patients with type 2 diabetes:A Genetics of Diabetes Audit and Research Study in Tayside Scotland (Go-DARTS) Study. Circ. Cardiovasc. Genet. 2009, 2, 255–259. [Google Scholar] [CrossRef] [Green Version]
- Freathy, R.M.; Timpson, N.J.; Lawlor, D.A.; Pouta, A.; Ben-Shlomo, Y.; Ruokonen, A.; Ebrahim, S.; Shields, B.; Zeggini, E.; Weedon, M.N.; et al. Common variation in the FTO gene alters diabetes-related metabolic traits to the extent expected given its effect on BMI. Diabetes 2008, 57, 1419–1426. [Google Scholar] [CrossRef] [Green Version]
- Jackson, K.G.; Poppitt, S.D.; Minihane, A.M. Postprandial lipemia and cardiovascular disease risk: Interrelationships between dietary, physiological and genetic determinants. Atherosclerosis 2012, 220, 22–33. [Google Scholar] [CrossRef]
- Zilversmit, D.B. Atherogenesis: A postprandial phenomenon. Circulation 1979, 60, 473–485. [Google Scholar] [CrossRef] [Green Version]
- Dias, C.B.; Moughan, P.J.; Wood, L.G.; Singh, H.; Garg, M.L. Postprandial lipemia: Factoring in lipemic response for ranking foods for their healthiness. Lipids Health Dis. 2017, 16, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bansal, S.; Buring, J.E.; Rifai, N.; Mora, S.; Sacks, F.M.; Ridker, P.M. Fasting compared with nonfasting triglycerides and risk of cardiovascular events in women. J. Am. Med. Assoc. 2007, 298, 309–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorling, J.L.; Clayton, D.J.; Jones, J.; Carter, W.G.; Thackray, A.E.; King, J.A.; Pucci, A.; Batterham, R.L.; Stensel, D.J. A randomized crossover trial assessing the effects of acute exercise on appetite, circulating ghrelin concentrations, and butyrylcholinesterase activity in normal-weight males with variants of the obesity-linked FTO rs9939609 polymorphism. Am. J. Clin. Nutr. 2019, 110, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Broom, D.R.; Stensel, D.J.; Bishop, N.C.; Burns, S.F.; Miyashita, M. Exercise-induced suppression of acylated ghrelin in humans. J. Appl. Physiol. 2007, 102, 2165–2171. [Google Scholar] [CrossRef]
- Schenk, S.; Horowitz, J.F. Acute exercise increases triglyceride synthesis in skeletal muscle and prevents fatty acid-induced insulin resistance. J. Clin. Investig. 2007, 117, 1690–1698. [Google Scholar] [CrossRef]
- Nestel, P.J.; Carroll, K.F.; Havenstein, N. Plasma triglyceride response to carbohydrates, fats and caloric intake. Metabolism 1970, 19, 1–18. [Google Scholar] [CrossRef]
- Robertson, M.D.; Henderson, R.; Vist, G.E.; Rumsey, R.D. Extended effects of evening meal carbohydrate-to-fat ratio on fasting and postprandial substrate metabolism. Am. J. Clin. Nutr. 2002, 75, 505–510. [Google Scholar] [CrossRef] [Green Version]
- Chandarana, K.; Drew, M.E.; Emmanuel, J.; Karra, E.; Gelegen, C.; Chan, P.; Cron, N.J.; Batterham, R.L. Subject standardization, acclimatization, and sample processing affect gut hormone levels and appetite in humans. Gastroenterology 2009, 136, 2115–2126. [Google Scholar] [CrossRef]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis model assessment: Insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences; Lawrence Erlbaum Associates: Hillsdale, NJ, USA, 1988; Volume 2, ISBN 0805802835. [Google Scholar]
- Faul, F.; Erdfelder, E.; Lang, A.G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]
- Hunt, S.C.; Stone, S.; Xin, Y.; Scherer, C.A.; Magness, C.L.; Iadonato, S.P.; Hopkins, P.N.; Adams, T.D. Association of the FTO gene with BMI. Obes. (Silver Spring Md.) 2008, 16, 902–904. [Google Scholar] [CrossRef] [PubMed]
- Hokanson, J.E.; Austin, M.A. Plasma triglyceride level is a risk factor for cardiovascular disease independent of high-density lipoprotein cholesterol level: A metaanalysis of population-based prospective studies. J. Cardiovasc. Risk 1996, 3, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, M.; Wessman, Y.; Almgren, P.; Groop, L. High levels of nonesterified fatty acids are associated with increased familial risk of cardiovascular disease. Arterioscler. Thromb. Vasc. Biol. 2000, 6, 1588–1594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cecil, J.E.; Tavendale, R.; Watt, P.; Hetherington, M.M.; Palmer, C.N.A. An obesity-associated FTO gene variant and increased energy intake in children. N. Engl. J. Med. 2008, 359, 2558–2566. [Google Scholar] [CrossRef]
- Ahmad, S.; Poveda, A.; Shungin, D.; Barroso, I.; Hallmans, G.; Renström, F.; Franks, P.W. Established BMI-associated genetic variants and their prospective associations with BMI and other cardiometabolic traits: The GLACIER Study. Int. J. Obes. 2016. [Google Scholar] [CrossRef]
- Zhao, T.-J.; Liang, G.; Li, R.L.; Xie, X.; Sleeman, M.W.; Murphy, A.J.; Valenzuela, D.M.; Yancopoulos, G.D.; Goldstein, J.L.; Brown, M.S. Ghrelin O-acyltransferase (GOAT) is essential for growth hormone-mediated survival of calorie-restricted mice. Proc. Natl. Acad. Sci. USA 2010, 107, 7467–7472. [Google Scholar] [CrossRef] [Green Version]
- Broglio, F.; Arvat, E.; Benso, A.; Gottero, C.; Muccioli, G.; Papotti, M.; Van Der Lely, A.J.; Deghenghi, R.; Ghigo, E. Ghrelin, a natural gh secretagogue produced by the stomach, induces hyperglycemia and reduces insulin secretion in humans. J. Clin. Endocrinol. Metab. 2001, 86, 5083–5086. [Google Scholar] [CrossRef]
- Moffitt, T.E.; Belsky, D.W.; Danese, A.; Poulton, R.; Caspi, A. The Longitudinal Study of Aging in Human Young Adults: Knowledge gaps and research agenda. J. Gerontol. Ser. A 2017, 72, 210–215. [Google Scholar] [CrossRef] [Green Version]

| AA (n =12) | TT (n = 12) | TT vs. AA Mean Difference (95% CI 1) | |
|---|---|---|---|
| Age (year) | 20.9 ± 3.5 | 21.3 ± 3.6 | −0.4 (−3.4 to 2.6) |
| Height (cm) | 181.6 ± 5.8 | 177.5 ± 6.5 | 4.1 (−1.2 to 9.3) |
| Weight (kg) | 77.6 ± 11.3 | 73.8 ± 6.9 | 3.9 (−4.1 to 11.8) |
| Body mass index (kg/m2) | 23.5 ± 2.7 | 23.5 ± 2.3 | 0.0 (−2.1 to 2.1) |
| Waist circumference (cm) | 80.3 ± 6.1 | 78.1 ± 4.1 | 2.2 (−2.2 to 6.6) |
| Body fat percentage (%) | 15.6 ± 5.1 | 13.9 ± 4.7 | 1.7 (−2.4 to 5.9) |
| Lean body mass (kg) | 65.2 ± 7.4 | 63.3 ± 4.2 | 1.9 (−3.2 to 7.0) |
| Peak oxygen uptake (mL/kg/minute) | 55.8 ± 5.8 | 56.6 ± 4.9 | −0.8 (−5.4 to 3.7) |
| Physical activity (metabolic equivalent minutes/week) | 4368 ± 1968 | 4790 ± 2728 | −423 (−2436 to 1591) |
| AA (n = 12) | TT (n = 12) | TT Vs. AA Mean Difference (95% CI 1) | Effect Size | |
|---|---|---|---|---|
| Triacylglycerol (mmol/L) | ||||
| Baseline | 0.77 ± 0.08 | 0.88 ± 0.07 | −0.11 (−0.32 to 0.10) | 0.44 |
| Pre-meal | 0.74 ± 0.08 | 0.81 ± 0.07 | −0.07 (−0.28 to 0.14) | 0.28 |
| Non-esterified fatty acids (mmol/L) | ||||
| Baseline | 0.46 ± 0.09 | 0.43 ± 0.06 | 0.03 (−0.18 to 0.25) | 0.12 |
| Pre-meal | 0.41 ± 0.06 | 0.45 ± 0.06 | −0.03 (−0.21 to 0.14) | 0.16 |
| Total cholesterol (mmol/L) | ||||
| Baseline | 3.40 ± 0.12 | 3.41 ± 0.18 | −0.01 (−0.47 to 0.45) | 0.02 |
| Low-density lipoprotein cholesterol (mmol/L) | ||||
| Baseline | 1.77 ± 0.12 | 1.83 ± 0.14 | −0.07 (−0.45 to 0.32) | 0.15 |
| High-density lipoprotein cholesterol (mmol/L) | ||||
| Baseline | 1.28 ± 0.07 | 1.24 ± 0.11 | 0.05 (−0.22 to 0.31) | 0.15 |
| Glucose (mmol/L) | ||||
| Baseline | 5.35 ± 0.11 | 5.26 ± 0.06 | 0.09 (−0.17 to 0.35) | 0.30 |
| Pre-meal | 5.18 ± 0.08 | 5.15 ± 0.04 | 0.03 (−0.16 to 0.23) | 0.14 |
| Insulin (pmol/L) | ||||
| Baseline | 3.34 ± 0.47 | 3.09 ± 0.52 | 0.25 (−1.21 to 1.72) | 0.15 |
| Pre-meal | 2.82 ± 0.34 | 3.26 ± 0.55 | −0.44 (−1.76 to 0.89) | 0.28 |
| HOMA-IR | ||||
| Baseline | 0.81 ± 0.12 | 0.72 ± 0.12 | 0.08 (−0.27 to 0.44) | 0.20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dorling, J.L.; Thackray, A.E.; King, J.A.; Pucci, A.; Goltz, F.R.; Batterham, R.L.; Stensel, D.J. No Influence of the Fat Mass and Obesity-Associated Gene rs9939609 Single Nucleotide Polymorphism on Blood Lipids in Young Males. Nutrients 2020, 12, 3857. https://doi.org/10.3390/nu12123857
Dorling JL, Thackray AE, King JA, Pucci A, Goltz FR, Batterham RL, Stensel DJ. No Influence of the Fat Mass and Obesity-Associated Gene rs9939609 Single Nucleotide Polymorphism on Blood Lipids in Young Males. Nutrients. 2020; 12(12):3857. https://doi.org/10.3390/nu12123857
Chicago/Turabian StyleDorling, James L., Alice E. Thackray, James A. King, Andrea Pucci, Fernanda R. Goltz, Rachel L. Batterham, and David J. Stensel. 2020. "No Influence of the Fat Mass and Obesity-Associated Gene rs9939609 Single Nucleotide Polymorphism on Blood Lipids in Young Males" Nutrients 12, no. 12: 3857. https://doi.org/10.3390/nu12123857





