In Vitro Evaluation of the Effects of Commercial Prebiotic GOS and FOS Products on Human Colonic Caco–2 Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Oligosaccharides
2.2. Cells and Experimental Design
2.3. Integrity of Tight Junctions
2.4. RNA Extraction and Qualification
2.5. Library Preparation for mRNA Sequencing
2.6. Quality Control and Mapping Analyses
2.7. Functional Analysis of Differentially Expressed Genes
2.8. Quantitative Real-Time PCR Validation
2.9. Statistical Analysis
2.10. Data Availability
3. Results
3.1. Influence of GOS and FOS on Epithelial Monolayer Tight Junction Integrity
3.2. Transcriptome Sequencing and Functional Annotation
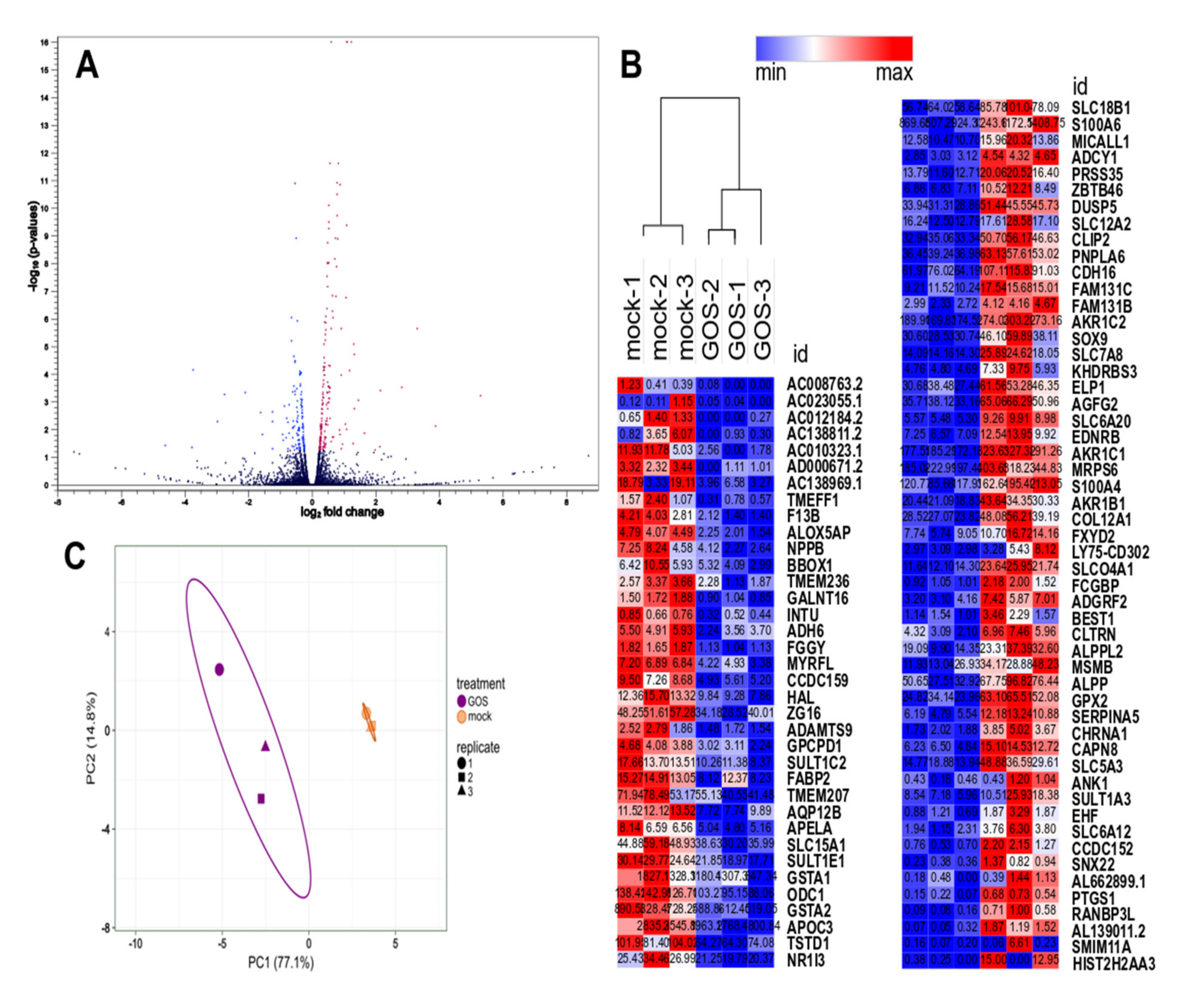
3.3. Analysis of Differential Gene Expression
3.4. Validation of Differentially Expressed Genes by RT-qPCR
3.5. Functional Analysis of Differentially Expressed Genes
4. Discussion
4.1. FOS Treatment Associated with 12 Differentially Expressed Genes
4.2. GOS Treatment Associated with 53 up-Regulated Genes
4.3. GOS Treatment Associated with 36 Down-Regulated Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. J. Nutr. 1995, 125, 1401–1412. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, S.; Macfarlane, G.T.; Cummings, J.H. Review article: Prebiotics in the gastrointestinal tract. Aliment. Pharmacol. Ther. 2006, 24, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Schley, P.D.; Field, C.J. The immune-enhancing effects of dietary fibres and prebiotics. Br. J. Nutr. 2002, 87, S221–S230. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, S.S.; Kuipers, B.J.H.; Dijkhuizen, L.; Kamerling, J.P. Comparative structural characterization of 7 commercial galacto-oligosaccharide (GOS) products. Carbohydr. Res. 2016, 425, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Martins, G.N.; Ureta, M.M.; Tymczyszyn, E.E.; Castilho, P.C.; Gomez-Zavaglia, A. Technological aspects of the production of fructo and galacto-oligosaccharides. Enzymatic synthesis and hydrolysis. Front. Nutr. 2019, 6, 78. [Google Scholar] [CrossRef]
- Niness, K.R. Inulin and Oligofructose: What are they? J. Nutr. 1999, 129, S1402–S1406. [Google Scholar] [CrossRef]
- Paganini, D.; Uyoga, M.A.; Cercamondi, C.I.; Moretti, D.; Mwasi, E.; Schwab, C.; Bechtler, S.; Mutuku, F.M.; Galetti, V.; Lacroix, C.; et al. Consumption of galacto-oligosaccharides increases iron absorption from a micronutrient powder containing ferrous fumarate and sodium iron EDTA: A stable-isotope study in Kenyan infants. Am. J. Clin. Nutr. 2017, 106, 1020–1031. [Google Scholar] [CrossRef]
- Hughes, C.; Davoodi-Semiromi, Y.; Colee, J.C.; Culpepper, T.; Dahl, W.J.; Mai, V.; Christman, M.C.; Langkamp-Henken, B. Galactooligosaccharide supplementation reduces stress-induced gastrointestinal dysfunction and days of cold or flu: A randomized, double-blind, controlled trial in healthy university students. Am. J. Clin. Nutr. 2011, 93, 1305–1311. [Google Scholar] [CrossRef]
- Schmidt, K.; Cowen, P.J.; Harmer, C.J.; Tzortzis, G.; Errington, S.; Burnet, P.W.J. Prebiotic intake reduces the waking cortisol response and alters emotional bias in healthy volunteers. Psychopharmacology (Berlin) 2015, 232, 1793–1801. [Google Scholar] [CrossRef]
- Liu, F.; Li, P.; Chen, M.; Luo, Y.; Prabhakar, M.; Zheng, H.; He, Y.; Qi, Q.; Long, H.; Zhang, Y.; et al. Fructooligosaccharide (FOS) and galactooligosaccharide (GOS) increase Bifidobacterium but reduce butyrate producing bacteria with adverse glycemic metabolism in healthy young population. Sci. Rep. 2017, 7, 11789. [Google Scholar] [CrossRef] [PubMed]
- Richards, P.J.; Flaujac Lafontaine, G.M.; Connerton, P.L.; Liang, L.; Asiani, K.; Fish, N.M.; Connerton, I.F. Galacto-oligosaccharides modulate the juvenile gut microbiome and innate immunity to improve broiler chicken performance. mSystems 2020, 5, e00827-19. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, S.; Zaneb, H.; Masood, S.; Yousaf, S.; Usman, M.M.; Rehman, H.F.; Sikandar, A.; Rehman, H. Influence of β-galacto-oligosaccharide on growth performance and components of intestinal barrier in broilers during heat stress. S. Afr. J. Anim. Sci. 2017, 47, 616–625. [Google Scholar] [CrossRef][Green Version]
- Varasteh, S.; Braber, S.; Akbari, P.; Garssen, J.; Fink-Gremmels, J. Differences in susceptibility to heat stress along the chicken intestine and the protective effects of galacto- oligosaccharides. PLoS ONE 2015, 10, e0138975. [Google Scholar] [CrossRef]
- Flaujac Lafontaine, G.M.; Richards, P.J.; Connerton, P.L.; O’Kane, P.M.; Ghaffar, N.M.; Cummings, N.J.; Fish, N.M.; Connerton, I.F. Prebiotic driven increases in IL-17A do not prevent Campylobacter jejuni colonization of chickens. Front. Microbiol. 2020, 10, 3033. [Google Scholar] [CrossRef]
- Alizadeh, A.; Akbari, P.; Difilippo, E.; Schols, H.A.; Ulfman, L.H.; Schoterman, M.H.C.; Garssen, J.; Fink-Gremmels, J.; Braber, S. The piglet as a model for studying dietary components in infant diets: Effects of galacto-oligosaccharides on intestinal functions. Br. J. Nutr. 2016, 115, 605–618. [Google Scholar] [CrossRef]
- Tian, S.; Wang, J.; Yu, H.; Wang, J.; Zhu, W. Changes in ileal microbial composition and microbial metabolism by an early-life galacto-oligosaccharides intervention in a neonatal porcine model. Nutrients 2019, 11, 1753. [Google Scholar] [CrossRef]
- Le Dréan, G.; Pocheron, A.L.; Billard, H.; Grit, I.; Pagniez, A.; Parnet, P.; Chappuis, E.; Rolli-Derkinderen, M.; Michel, C. Neonatal consumption of oligosaccharides greatly increases L-cell density without significant consequence for adult eating behavior. Nutrients 2019, 11, 1967. [Google Scholar] [CrossRef]
- Perdijk, O.; Van Baarlen, P.; Fernandez-Gutierrez, M.M.; Van Den Brink, E.; Schuren, F.H.J.; Brugman, S.; Savelkoul, H.F.J.; Kleerebezem, M.; Neerven, R.J.J. Van Sialyllactose and galactooligosaccharides promote epithelial barrier functioning and distinctly modulate microbiota composition and short chain fatty acid production in vitro. Front. Immunol. 2019, 10, 94. [Google Scholar] [CrossRef]
- Akbari, P.; Braber, S.; Alizadeh, A.; Verheijden, K.A.; Schoterman, M.H.; Kraneveld, A.D.; Garssen, J.; Fink-Gremmels, J. Galacto-oligosaccharides protect the intestinal barrier by maintaining the tight junction network and modulating the inflammatory responses after a challenge with the mycotoxin deoxynivalenol in Human Caco-2 cell monolayers and B6C3F1 mice. J. Nutr. 2015, 145, 1604–1613. [Google Scholar] [CrossRef]
- Searle, L.E.J.; Cooley, W.A.; Jones, G.; Nunez, A.; Crudgington, B.; Weyer, U.; Dugdale, A.H.; Tzortzis, G.; Collins, J.W.; Woodward, M.J.; et al. Purified galactooligosaccharide, derived from a mixture produced by the enzymic activity of Bifidobacterium bifidum, reduces Salmonella enterica serovar Typhimurium adhesion and invasion in vitro and in vivo. J. Med. Microbiol. 2010, 59, 1428–1439. [Google Scholar] [CrossRef]
- Whisner, C.M.; Martin, B.R.; Schoterman, M.H.C.; Nakatsu, C.H.; McCabe, L.D.; McCabe, G.P.; Wastney, M.E.; van den Heuvel, E.G.H.M.; Weaver, C.M. Galacto-oligosaccharides increase calcium absorption and gut bifidobacteria in young girls: A double-blind cross-over trial. Br. J. Nutr. 2013, 110, 1292–1303. [Google Scholar] [CrossRef]
- Van Den Heuvel, E.G.H.M.; Muys, T.; Van Dokkum, W.; Schaafsma, G. Oligofructose stimulates calcium absorption in adolescents. Am. J. Clin. Nutr. 1999, 69, 544–548. [Google Scholar] [CrossRef] [PubMed]
- Zafar, T.A.; Weaver, C.M.; Zhao, Y.; Martin, B.R.; Wastney, M.E. Nondigestible oligosaccharides increase calcium absorption and suppress bone resorption in ovariectomized rats. J. Nutr. 2004, 134, 399–402. [Google Scholar] [CrossRef] [PubMed]
- Fransen, F.; Sahasrabudhe, N.M.; Elderman, M.; Bosveld, M.; El Aidy, S.; Hugenholtz, F.; Borghuis, T.; Kousemaker, B.; Winkel, S.; van der Gaast-de Jongh, C.; et al. β2→1-fructans modulate the immune system in vivo in a microbiota-dependent and -independent fashion. Front. Immunol. 2017, 8, 16. [Google Scholar] [CrossRef]
- Babu, U.S.; Sommers, K.; Harrison, L.M.; Balan, K.V. Effects of fructooligosaccharide-inulin on Salmonella-killing and inflammatory gene expression in chicken macrophages. Vet. Immunol. Immunopathol. 2012, 149, 92–96. [Google Scholar] [CrossRef]
- Srinivasan, B.; Kolli, A.R.; Esch, M.B.; Abaci, H.E.; Shuler, M.L.; Hickman, J.J. TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. 2015, 20, 107–126. [Google Scholar] [CrossRef]
- Natoli, M.; Zucco, F.; Felsani, A.; Leoni, B.D.; D’Agnano, I. Good Caco-2 cell culture practices. Toxicol. Vitr. 2012, 26, 1243–1246. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Minguet, E.G.; Segard, S.; Charavay, C.; Parcy, F. MORPHEUS, a webtool for transcription factor binding analysis using position weight matrices with dependency. PLoS ONE 2015, 10, e0135586. [Google Scholar] [CrossRef] [PubMed]
- Heberle, H.; Meirelles, V.G.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Carbon, S.; Douglass, E.; Dunn, N.; Good, B.; Harris, N.L.; Lewis, S.E.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; et al. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 2005, 21, 3439–3440. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/ Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Luo, W.; Friedman, M.S.; Shedden, K.; Hankenson, K.D.; Woolf, P.J. GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinform. 2009, 10, 161. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Connerton, P.L.; Richards, P.J.; Lafontaine, G.M.; O’Kane, P.M.; Ghaffar, N.; Cummings, N.J.; Smith, D.L.; Fish, N.M.; Connerton, I.F. The effect of the timing of exposure to Campylobacter jejuni on the gut microbiome and inflammatory responses of broiler chickens. Microbiome 2018, 6, 88. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2002, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Larionov, A.; Krause, A.; Miller, W.R. A standard curve based method for relative real time PCR data processing. BMC Bioinform. 2005, 6, 62. [Google Scholar] [CrossRef]
- Baggerly, K.A.; Deng, L.; Morris, J.S.; Aldaz, C.M. Differential expression in SAGE: Accounting for normal between-library variation. Bioinformatics 2003, 19, 1477–1483. [Google Scholar] [CrossRef]
- Rivals, I.; Personnaz, L.; Taing, L.; Potier, M.C. Enrichment or depletion of a GO category within a class of genes: Which test? Bioinformatics 2007, 23, 401–407. [Google Scholar] [CrossRef]
- Roberfroid, M.; Gibson, G.R.; Hoyles, L.; McCartney, A.L.; Rastall, R.; Rowland, I.; Wolvers, D.; Watzl, B.; Szajewska, H.; Stahl, B.; et al. Prebiotic effects: Metabolic and health benefits. Br. J. Nutr. 2010, 104, S1–S63. [Google Scholar] [CrossRef]
- Volpe, D.A.; Faustino, P.J.; Ciavarella, A.B.; Asafu-Adjaye, E.B.; Ellison, C.D.; Yu, L.X.; Hussain, A.S. Classification of drug permeability with a Caco-2 cell monolayer assay. Clin. Res. Regul. Aff. 2007, 24, 39–47. [Google Scholar] [CrossRef]
- Keemink, J.; Bergström, C.A.S. Caco-2 cell conditions enabling studies of drug absorption from digestible lipid-based formulations. Pharm. Res. 2018, 35, 74. [Google Scholar] [CrossRef]
- Wu, R.Y.; Abdullah, M.; Määttänen, P.; Pilar, A.V.C.; Scruten, E.; Johnson-Henry, K.C.; Napper, S.; O’Brien, C.; Jones, N.L.; Sherman, P.M. Protein kinase Cσ signaling is required for dietary prebiotic-induced strengthening of intestinal epithelial barrier function. Sci. Rep. 2017, 7, 40820. [Google Scholar] [CrossRef]
- Shoaf, K.; Mulvey, G.L.; Armstrong, G.D.; Hutkins, R.W. Prebiotic galactooligosaccharides reduce adherence of enteropathogenic Escherichia coli to tissue culture cells. Infect. Immun. 2006, 74, 6920–6928. [Google Scholar] [CrossRef] [PubMed]
- Putt, K.K.; Pei, R.; White, H.M.; Bolling, B.W. Yogurt inhibits intestinal barrier dysfunction in Caco-2 cells by increasing tight junctions. Food Funct. 2017, 8, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Akbari, P.; Braber, S.; Gremmels, H.; Koelink, P.J.; Verheijden, K.A.T.; Garssen, J.; Fink-Gremmels, J. Deoxynivalenol: A trigger for intestinal integrity breakdown. FASEB J. 2014, 28, 2414–2429. [Google Scholar] [CrossRef] [PubMed]
- Dalman, M.R.; Deeter, A.; Nimishakavi, G.; Duan, Z.H. Fold change and p-value cutoffs significantly alter microarray interpretations. BMC Bioinform. 2012, 3, S11. [Google Scholar] [CrossRef]
- Mccarthy, D.J.; Smyth, G.K. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics 2009, 25, 765–771. [Google Scholar] [CrossRef]
- Schurch, N.J.; Schofield, P.; Gierliński, M.; Cole, C.; Sherstnev, A.; Singh, V.; Wrobel, N.; Gharbi, K.; Simpson, G.G.; Owen-Hughes, T.; et al. How many biological replicates are needed in an RNA-Seq experiment and which differential expression tool should you use? RNA 2016, 22, 839–851. [Google Scholar] [CrossRef]
- St Laurent, G.; Shtokalo, D.; Tackett, M.R.; Yang, Z.; Vyatkin, Y.; Milos, P.M.; Seilheimer, B.; McCaffrey, T.A.; Kapranov, P. On the importance of small changes in RNA expression. Methods 2013, 63, 18–24. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef]
- Badawi, A.F.; Cavalieri, E.L.; Rogan, E.G. Role of human cytochrome P450 1A1, 1A2, 1B1, and 3A4 in the 2-, 4-, and 16α-hydroxylation of 17β-estradiol. Metabolism 2001, 50, 1001–1003. [Google Scholar] [CrossRef]
- Lucas, D.; Goulitquer, S.; Marienhagen, J.; Fer, M.; Dreano, Y.; Schwaneberg, U.; Amet, Y.; Corcos, L. Stereoselective epoxidation of the last double bond of polyunsaturated fatty acids by human cytochromes P450. J. Lipid Res. 2010, 51, 1125–1133. [Google Scholar] [CrossRef]
- Clayton, E.L.; Minogue, S.; Waugh, M.G. Mammalian phosphatidylinositol 4-kinases as modulators of membrane trafficking and lipid signaling networks. Prog. Lipid Res. 2013, 52, 294–304. [Google Scholar] [CrossRef] [PubMed]
- Wenz, J.J. Predicting the effect of steroids on membrane biophysical properties based on the molecular structure. Biochim. Biophys. Acta Biomembr. 2012, 1818, 896–906. [Google Scholar] [CrossRef] [PubMed]
- Vreeburg, R.A.M.; Bastiaan-Net, S.; Mes, J.J. Normalization genes for quantitative RT-PCR in differentiated Caco-2 cells used for food exposure studies. Food Funct. 2011, 2, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Colón, G.J.; Moore, B.B. Prostaglandin E2 as a regulator of immunity to pathogens. Pharmacol. Ther. 2018, 185, 135–146. [Google Scholar] [CrossRef]
- Hata, S.; Koyama, S.; Kawahara, H.; Doi, N.; Maeda, T.; Toyama-Sorimachi, N.; Abe, K.; Suzuki, K.; Sorimachi, H. Stomach-specific calpain, nCL-2, localizes in mucus cells and proteolyzes the β-subunit of coatomer complex, β-COP. J. Biol. Chem. 2006, 281, 11214–11224. [Google Scholar] [CrossRef]
- Morford, L.A.; Forrest, K.; Logan, B.; Overstreet, L.K.; Goebel, J.; Brooks, W.H.; Roszman, T.L. Calpain II colocalizes with detergent-insoluble rafts on human and Jurkat T-cells. Biochem. Biophys. Res. Commun. 2002, 295, 540–546. [Google Scholar] [CrossRef]
- Hood, J.L.; Brooks, W.H.; Rossman, T.L. Differential compartmentalization of the calpain/calpastatin network with the endoplasmic reticulum and Golgi apparatus. J. Biol. Chem. 2004, 279, 43126–43135. [Google Scholar] [CrossRef]
- Zhao, W.C.; Zhu, J.X.; Zhang, G.H.; Wong, C.H.; Chung, Y.W.; Chan, H.C. Effect of sodium ferulate on human colonic anion secretion and the underlying signaling mechanism. Biol. Pharm. Bull. 2005, 28, 1608–1611. [Google Scholar] [CrossRef][Green Version]
- Kliewer, S.A.; Sundseth, S.S.; Jones, S.A.; Brown, P.J.; Wisely, G.B.; Koble, C.S.; Devchand, P.; Wahli, W.; Willson, T.M.; Lenhard, J.M.; et al. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ. Proc. Natl. Acad. Sci. USA 1997, 94, 4318–4323. [Google Scholar] [CrossRef]
- Paumi, C.M.; Smitherman, P.K.; Townsend, A.J.; Morrow, C.S. Glutathione S-Transferases (GSTs) inhibit transcriptional activation by the Peroxisomal Proliferator-Activated Receptor γ (PPARγ) ligand, 15-deoxy-Δ12,14prostaglandin J2 (15-d-PGJ 2). Biochemistry 2004, 43, 2345–2352. [Google Scholar] [CrossRef]
- Hayes, J.D.; Flanagan, J.U.; Jowsey, I.R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 51–88. [Google Scholar] [CrossRef] [PubMed]
- Greene, E.R.; Huang, S.; Serhan, C.N.; Panigrahy, D. Regulation of inflammation in cancer by eicosanoids. Prostaglandins Other Lipid Mediat. 2011, 96, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Pidgeon, G.P.; Lysaght, J.; Krishnamoorthy, S.; Reynolds, J.V.; O’Byrne, K.; Nie, D.; Honn, K.V. Lipoxygenase metabolism: Roles in tumor progression and survival. Cancer Metastasis Rev. 2007, 26, 503–524. [Google Scholar] [CrossRef] [PubMed]
- Qamar, A.; Khetarpal, S.A.; Khera, A.V.; Qasim, A.; Rader, D.J.; Reilly, M.P. Plasma apolipoprotein C-III levels, triglycerides, and coronary artery calcification in type 2 diabetics. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1880–1888. [Google Scholar] [CrossRef]
- Khetarpal, S.A.; Zeng, X.; Millar, J.S.; Vitali, C.; Somasundara, A.V.H.; Zanoni, P.; Landro, J.A.; Barucci, N.; Zavadoski, W.J.; Sun, Z.; et al. A human APOC3 missense variant and monoclonal antibody accelerate apoC-III clearance and lower triglyceride-rich lipoprotein levels. Nat. Med. 2017, 23, 1086–1094. [Google Scholar] [CrossRef]
- Hirabayashi, J.; Hashidate, T.; Arata, Y.; Nishi, N.; Nakamura, T.; Hirashima, M.; Urashima, T.; Oka, T.; Futai, M.; Muller, W.E.; et al. Oligosaccharide specificity of galectins: A search by frontal affinity chromatography. Biochim. Biophys. Acta 2002, 1572, 232–254. [Google Scholar] [CrossRef]
- Bergström, J.H.; Birchenough, G.M.H.; Katona, G.; Schroeder, B.O.; Schütte, A.; Ermund, A.; Johansson, M.E.V.; Hansson, G.C. Gram-positive bacteria are held at a distance in the colon mucus by the lectin-like protein ZG16. Proc. Natl. Acad. Sci. USA 2016, 113, 13833–13838. [Google Scholar] [CrossRef]
- Meng, H.; Li, W.; Boardman, L.A.; Wang, L. Loss of ZG16 is associated with molecular and clinicopathological phenotypes of colorectal cancer. BMC Cancer 2018, 18, 433. [Google Scholar] [CrossRef]
- Wang, L.W.; Nandadasa, S.; Annis, D.S.; Dubail, X.J.; Mosher, D.F.; Willard, B.B.; Apte, S.S. A disintegrin-like and metalloproteinase domain with thrombospondin type 1 motif 9 (ADAMTS9) regulates fibronectin fibrillogenesis and turnover. J. Biol. Chem. 2019, 294, 9924–9936. [Google Scholar] [CrossRef]
- Du, W.; Wang, S.; Zhou, Q.; Li, X.; Chu, J.; Chang, Z.; Tao, Q.; Ng, E.K.O.; Fang, J.; Sung, J.J.Y.; et al. ADAMTS9 is a functional tumor suppressor through inhibiting AKT/mTOR pathway and associated with poor survival in gastric cancer. Oncogene 2013, 32, 3319–3328. [Google Scholar] [CrossRef]
- Fei, F.; Qu, J.; Zhang, M.; Li, Y.; Zhang, S. S100A4 in cancer progression and metastasis: A systematic review. Oncotarget 2017, 8, 73219–73239. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.B.; Holmgren, J.; Larena, M.; Terrinoni, M.; Fang, Y.; Bresnick, A.R.; Xiang, Z. Deficiency in calcium-binding protein S100A4 impairs the adjuvant action of cholera toxin. Front. Immunol. 2017, 8, 1119. [Google Scholar] [CrossRef] [PubMed]
- Büchau, A.S.; Hassan, M.; Kukova, G.; Lewerenz, V.; Kellermann, S.; Würthner, J.U.; Wolf, R.; Walz, M.; Gallo, R.L.; Ruzicka, T. S100A15, an antimicrobial protein of the skin: Regulation by E. coli through toll-like receptor 4. J. Invest. Dermatol. 2007, 127, 2596–2604. [Google Scholar] [CrossRef]
- Yang, H.; Geiger, M. Cell penetrating SERPINA5 (Protein C inhibitor, PCI): More questions than answers. Semin. Cell Dev. Biol. 2017, 62, 187–193. [Google Scholar] [CrossRef]
- Malmström, E.; Mörgelin, M.; Malmsten, M.; Johansson, L.; Norrby-Teglund, A.; Shannon, O.; Schmidtchen, A.; Meijers, J.C.M.; Herwald, H. Protein C inhibitor-A novel antimicrobial agent. PLoS Pathog. 2009, 5, e1000698. [Google Scholar] [CrossRef]
- Akita, N.; Ma, N.; Okamoto, T.; Asanuma, K.; Yoshida, K.; Nishioka, J.; Shimaoka, M.; Suzuki, K.; Hayashi, T. Host protein C inhibitor inhibits tumor growth, but promotes tumor metastasis, which is closely correlated with hypercoagulability. Thromb. Res. 2015, 135, 1203–1208. [Google Scholar] [CrossRef]
- Chiquet, M.; Birk, D.E.; Bönnemann, C.G.; Koch, M. Collagen XII: Protecting bone and muscle integrity by organizing collagen fibrils. Int. J. Biochem. Cell Biol. 2014, 53, 51–54. [Google Scholar] [CrossRef]
- Hicks, D.; Farsani, G.T.; Laval, S.; Collins, J.; Sarkozy, A.; Martoni, E.; Shah, A.; Zou, Y.; Koch, M.; Bönnemann, C.G.; et al. Mutations in the collagen XII gene define a new form of extracellular matrix-related myopathy. Hum. Mol. Genet. 2014, 23, 2353–2363. [Google Scholar] [CrossRef]





| Composition | |
|---|---|
| Nutrabiotic® GOS | 75% (w/w) dry solids (ds) |
| Galacto-oligosaccharides | 66.5 (w/w ds) |
| Lactose | 10.1 (w/w ds) |
| Glucose | 21.8 (w/w ds) |
| Galactose | 1.6 (w/w ds) |
| Orafti® L95 FOS | 74.7% (w/w) dry solids |
| Fructo-oligosaccharides | 94.8 (w/w ds) |
| Fructose | 3 (w/w ds) |
| Sucrose | 2 (w/w ds) |
| Glucose | 0.2 (w/w ds) |
| Media | Basic media* supplementation |
|---|---|
| Cell growth & confluence | |
| + 10% (v/v) FBS | |
| + antibiotics 100 U/mL (penicillin, streptomycin) | |
| Conditioning & treatment | |
| GOS 2% | + 2% (v/v) Nutrabiotic® GOS |
| FOS 2% | + 2% (v/v) Orafti® L95 FOS |
| GOS mock | + 0.67 % (m/m) mono- and di-saccharides (glucose 4.796 g/L, galactose 0.352 g/L, lactose 2.126 g/L) |
| FOS mock | + 0.104 % (m/m) mono- and di-saccharides (glucose 0.044 g/L, fructose 0.660 g/L, sucrose 0.421 g/L) |
| Target Gene | Primer Sequence (5’–3’) | Product Size (bp) | NCBI Accession | RNA–Seq Identifier |
|---|---|---|---|---|
| CAPN8 | F: GGTCTAGGTGACTGCTGGCT R: AGCAGCTGTCCATTCTTGGT | 197 | NM_001143962.2 | ENSG00000203697 |
| COL12A1 | F: GGCAAGGCTATCCAGGTTCC R: TAAGCACGTGCGCAAACATC | 106 | NM_004370.6 | ENSG00000111799 |
| CYP1A1 | F: CCCCCACAGCACAACAAGAG R: GGGTGAGAAACCGTTCAGGT | 146 | NM_000499.5 | ENSG00000140465 |
| F13B | F: GGACACTTCCTCCTGAGTGTGT R: CGTCTGCAACAGCCCCATTC | 81 | NM_001994.3 | ENSG00000143278 |
| GALNT16 | F: CTGACCTTCGTGGAGGGTTC R: GGTCTGTCCGGGTCATCTTC | 84 | NM_020692.3 | ENSG00000100626 |
| GPX2 | F: TTTCAATACGTTCCGGGGCA R: CTGACAGTTCTCCTGATGTCCA | 169 | NM_002083 | ENSG00000176153 |
| RGPD5 | F: CAAGAAATTGCCTGTGCCCC R: TCCATCGAGGTGGTGTTTCG | 215 | NM_005054.3 | ENSG00000015568 |
| SLC5A3 | F: ATGCAGCGGGGTTGGTACA R: AGCAACACAGCAGGGTCAAA | 235 | NM_006933.7 | ENSG00000198743 |
| SULT1A3 | F: CGGTCTCCTACTACCATTTCC R: AGGACCCGTAGGACACTTC | 108 | NM_177552.3 | ENSG00000261052 |
| ACTB | F: CTGGAACGGTGAAGGTGACA R: AAGGGACTTCCTGTAACAATGCA | 140 | NM_001101.5 | |
| GAPDH | F: GGAGTCCACTGGCGTCTTCAC R: GAGGCATTGCTGATGATCTTGAGG | 165 | NM_002046.7 | |
| PUM1 | F: TGAGGTGTGCACCATGAAC R: CAGAATGTGCTTGCCATAGG | 187 | NM_014676.2 |
| Raw Read Count | Ignored Reads (Wrong Strand) | Reads Paired and Mapped | Fragments Mapped to Genes | Fragments Mapped as Intergenic | Protein Coding Genes | ||
|---|---|---|---|---|---|---|---|
| FOS experiment | mock1 | 47,141,326 | 615,415 (1.30%) | 88.25% | 95.87% | 4.13% | 96.47% |
| mock2 | 49,710,422 | 771,068 (1.55%) | 87.24% | 95.81% | 4.19% | 96.54% | |
| mock3 | 44,983,458 | 717,544 (1.59%) | 89.62% | 95.86% | 4.14% | 96.64% | |
| FOS1 | 49,264,466 | 842,951 (1.71%) | 85.56% | 95.47% | 4.53% | 96.35% | |
| FOS2 | 51,905,580 | 749,101 (1.44%) | 86.80% | 95.64% | 4.36% | 96.46% | |
| FOS3 | 49,519,788 | 688,065 (1.39%) | 89.41% | 95.46% | 4.54% | 96.59% | |
| GOS experiment | mock1 | 62,474,912 | 819,911 (1.31%) | 89.96% | 95.73% | 4.27% | 96.47% |
| mock2 | 45,810,896 | 589,565 (1.29%) | 91.01% | 95.86% | 4.14% | 96.59% | |
| mock3 | 49,459,368 | 649,851 (1.31%) | 88.43% | 95.88% | 4.12% | 96.48% | |
| GOS1 | 134,054,024 | 2,460,936 (1.83%) | 89.29% | 95.78% | 4.22% | 96.54% | |
| GOS2 | 59,524,110 | 1,153,619 (1.94%) | 87.68% | 95.87% | 4.13% | 96.49% | |
| GOS3 | 56,118,898 | 809,112 (1.44%) | 90.74% | 95.53% | 4.47% | 96.61% |
| KEGG Pathway | FDR q.val | Enriched Gene | Protein Coding Alias | Fold Change (mRNAseq) | FDR p–Value (mRNAseq) |
|---|---|---|---|---|---|
| GOS (UP) | |||||
| hsa00790 Folate biosynthesis | 0.0003 | AKR1B1 | Aldo-Keto Reductase | 1.8 | 1 × 10−11 |
| ALPP | Intestinal Alkaline Phosphatase | 2.1 | 4 × 10−10 | ||
| ALPG | Placental-Like Alkaline Phosphatase | 2.1 | 2 × 10−7 | ||
| hsa00730 Thiamine metabolism | 0.0025 | ALPP | |||
| ALPG | |||||
| hsa04918 Thyroid hormone synthesis | 0.0170 | GPX2 | Glutathione Peroxidase-Gastrointestinal | 2.1 | <1 × 10−12 |
| ADCY1 | Ca2+/Calmodulin-Activated Adenylyl Cyclase | 1.5 | 5 × 10−4 | ||
| FXYD2 | Sodium/Potassium-Transporting ATPase Subunit Gamma | 1.8 | 0.010 | ||
| hsa04970 Salivary secretion | 0.0359 | SLC12A2 | Basolateral Na-K-Cl Symporter | 1.5 | 0.004 |
| ADCY1 | |||||
| FXYD2 | |||||
| hsa04974 Protein digestion and absorption | 0.0359 | SLC7A8 | L-Type Amino Acid Transporter 2 | 1.6 | 9 × 10−7 |
| COL12A1 | Collagen Type XII Alpha 1 Chain | 1.8 | 2 × 10−12 | ||
| FXYD2 | |||||
| hsa04972 Pancreatic secretion | 0.0495 | SLC12A2 | |||
| ADCY1 | |||||
| FXYD2 | |||||
| GOS (DOWN) | |||||
| hsa00480 Glutathione metabolism | 0.002 | ODC1 | Ornithine Decarboxylase 1 | −1.5 | 1 × 10−11 |
| GSTA2 | Glutathione S-Transferase Alpha 2 | −1.5 | 4 × 10−5 | ||
| GSTA1 | Glutathione S-Transferase Alpha 1 | −1.5 | 0.008 | ||
| hsa00982 Drug metabolism - cytochrome P | 0.005 | GSTA2 | |||
| hsa00980 Metabolism of xenobiotics by cytochrome P | 0.007 | GSTA1 | |||
| hsa05204 Chemical carcinogenesis | 0.009 | ADH6 | Alcohol Dehydrogenase 6 (Class V) | −1.7 | 0.004 |
| FOS (UP) | |||||
| hsa04913 Ovarian steroidogenesis | 0.0001 | CYP1A1 | Cytochrome P450 Family 1 Subfamily A Member 1 (Aryl Hydrocarbon Hydroxylase) | 1.8 | <1 × 10−12 |
| PLA2G4B | Phospholipase A2 Group IVB | 3.7 | 0.008 | ||
| hsa00590 Arachidonic acid metabolism | 0.0002 | PLA2G4B | |||
| GPX2 | Gastrointestinal Glutathione Peroxidase | 1.5 | 0.002 | ||
| hsa01100 Metabolic pathways | 0.0130 | NDUFC2-KCTD14 | NDUFC2-KCTD14 Readthrough Transcript Protein (NADH Dehydrogenase (Ubiquinone) 1 Subunit C2, Isoform 2) | 3.7 | 0.039 |
| AC104662.2 | predicted type II PI4 kinase protein family (PI4K2B) | 28.2 | 0.009 | ||
| PLA2G4B | |||||
| CYP1A1 |
| GO Cell Compartment Enrichment | FDR q.val | Enriched Gene | Protein Coding Alias | Fold Change (mRNAseq) | FDR p–Value (mRNAseq ) |
|---|---|---|---|---|---|
| GOS | |||||
| GO:0062023 Collagen-containing extracellular matrix | 0.018 | ZG16 | Zymogen Granule Protein 16 (Jacalin-Like Lectin Domain Containing) | −1.6 | 9 × 10−7 |
| APOC3 | Apolipoprotein C3 | −1.5 | 3 × 10−5 | ||
| ADAMTS9 | A Disintegrin And Metalloproteinase with ThromboSpondin Motifs 9 | −1.5 | 4 × 10−2 | ||
| SERPINA5 | Serine (Or Cysteine) Proteinase Inhibitor, clade A (Alpha-1 Antiproteinase, Antitrypsin), Member 5 | 2.1 | <1 × 10−12 | ||
| COL12A1 | Collagen type XII Proteoglycan | 1.8 | 2 × 10−12 | ||
| S100A4 | S100 Calcium Binding Protein A4 | 1.7 | 2 × 10−10 | ||
| S100A6 | S100 Calcium-Binding Protein A6 (Calcyclin) | 1.5 | 5 × 10−5 | ||
| FOS | |||||
| GO:0005743 Mitochondrial inner membrane | 0.002 | CYP1A1 | Cytochrome P450 Family 1 Subfamily A Member 1 (Aryl Hydrocarbon Hydroxylase) | 1.8 | <1 × 10−12 |
| PLA2G4B | Phospholipase A2 Group IVB | 3.7 | 0.008 | ||
| NDUFC2-KCTD14 | NDUFC2-KCTD14 Readthrough Transcript Protein (NADH Dehydrogenase (Ubiquinone) 1 Subunit C2, Isoform 2) | 3.7 | 0.039 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lafontaine, G.M.F.; Fish, N.M.; Connerton, I.F. In Vitro Evaluation of the Effects of Commercial Prebiotic GOS and FOS Products on Human Colonic Caco–2 Cells. Nutrients 2020, 12, 1281. https://doi.org/10.3390/nu12051281
Lafontaine GMF, Fish NM, Connerton IF. In Vitro Evaluation of the Effects of Commercial Prebiotic GOS and FOS Products on Human Colonic Caco–2 Cells. Nutrients. 2020; 12(5):1281. https://doi.org/10.3390/nu12051281
Chicago/Turabian StyleLafontaine, Geraldine M. Flaujac, Neville M. Fish, and Ian F. Connerton. 2020. "In Vitro Evaluation of the Effects of Commercial Prebiotic GOS and FOS Products on Human Colonic Caco–2 Cells" Nutrients 12, no. 5: 1281. https://doi.org/10.3390/nu12051281
APA StyleLafontaine, G. M. F., Fish, N. M., & Connerton, I. F. (2020). In Vitro Evaluation of the Effects of Commercial Prebiotic GOS and FOS Products on Human Colonic Caco–2 Cells. Nutrients, 12(5), 1281. https://doi.org/10.3390/nu12051281






