The Many Ages of Microbiome–Gut–Brain Axis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice and Behavioral Tests
2.2. Bacterial DNA Extraction and 16s rRNA Sequencing
2.3. Illumina Data Processing and Microbiota Characterization
2.4. Data Analysis and Statistics
3. Results
3.1. Sequencing Data Results
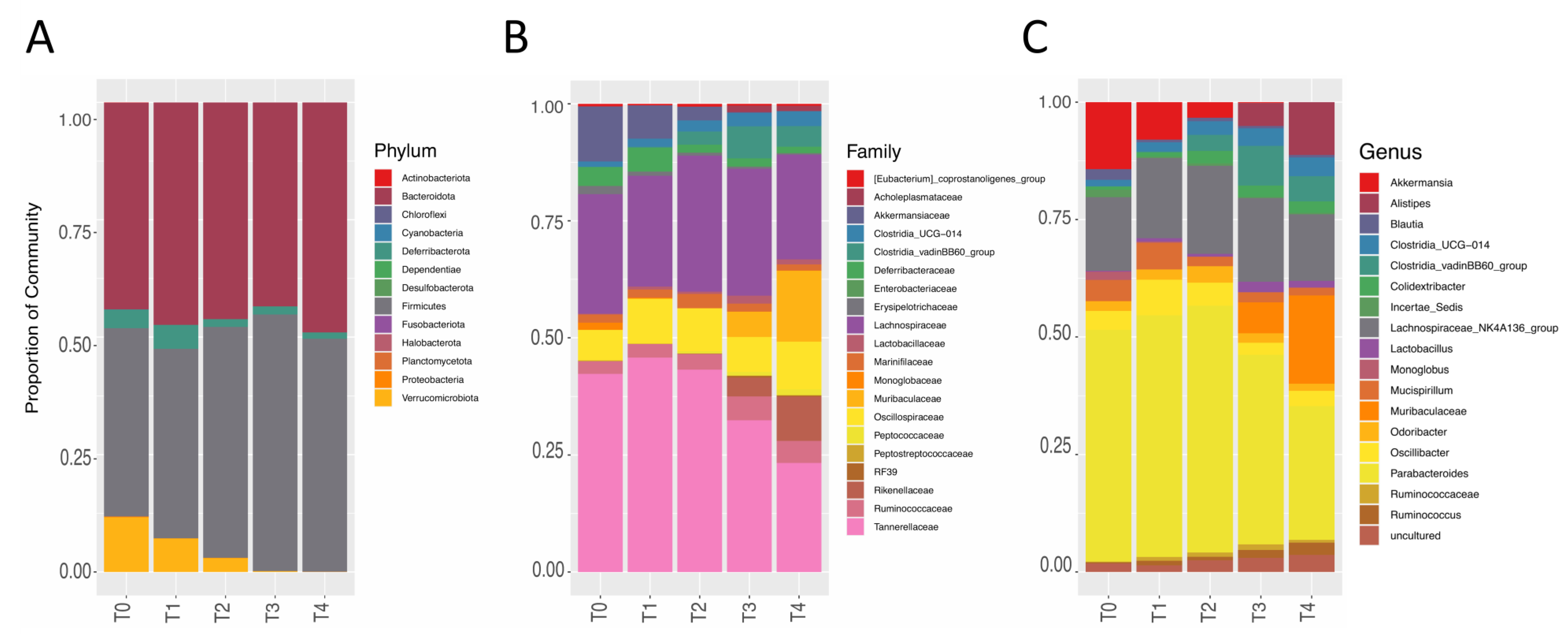
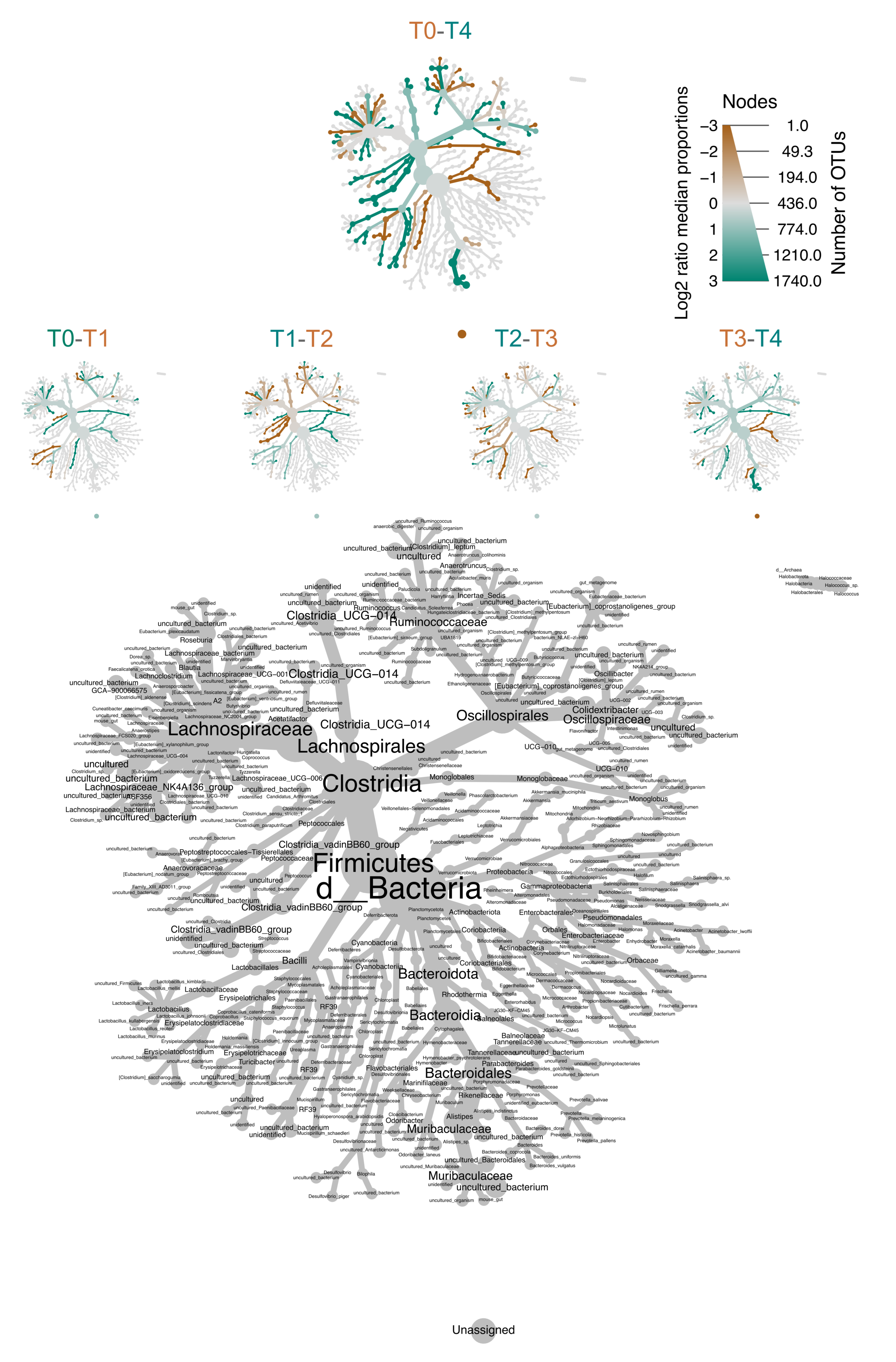
3.2. Aging Affects Overall Gut Microbiome Composition
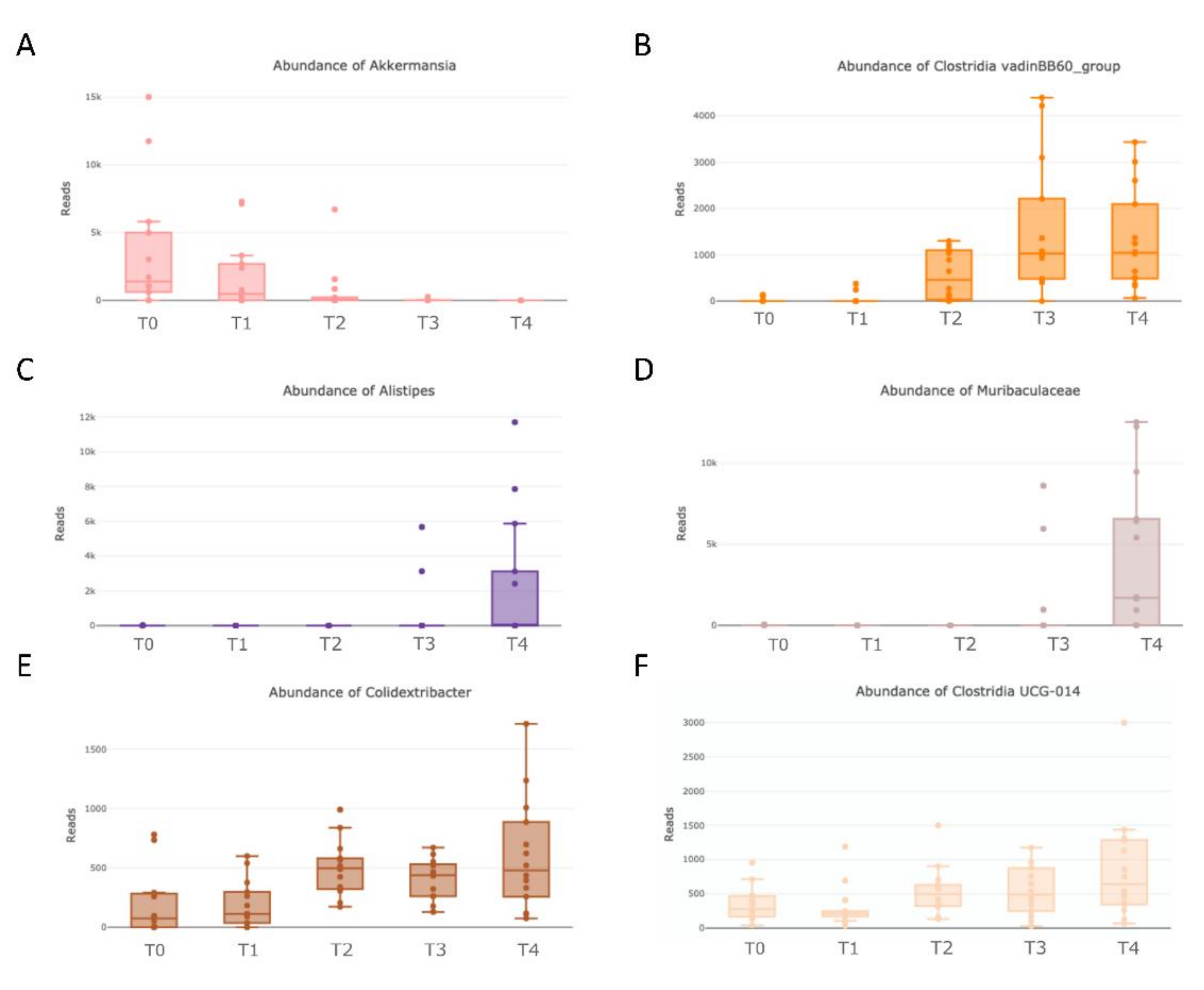
3.3. Detailed Microbiome Composition and Aging Distribution
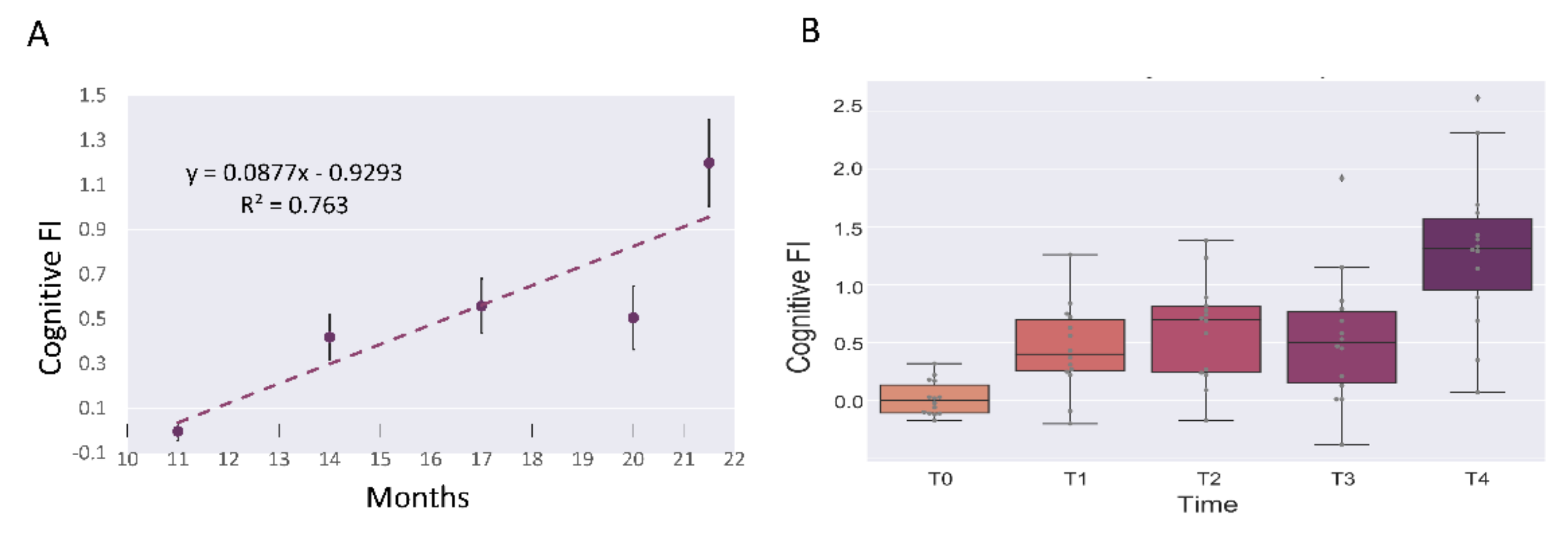
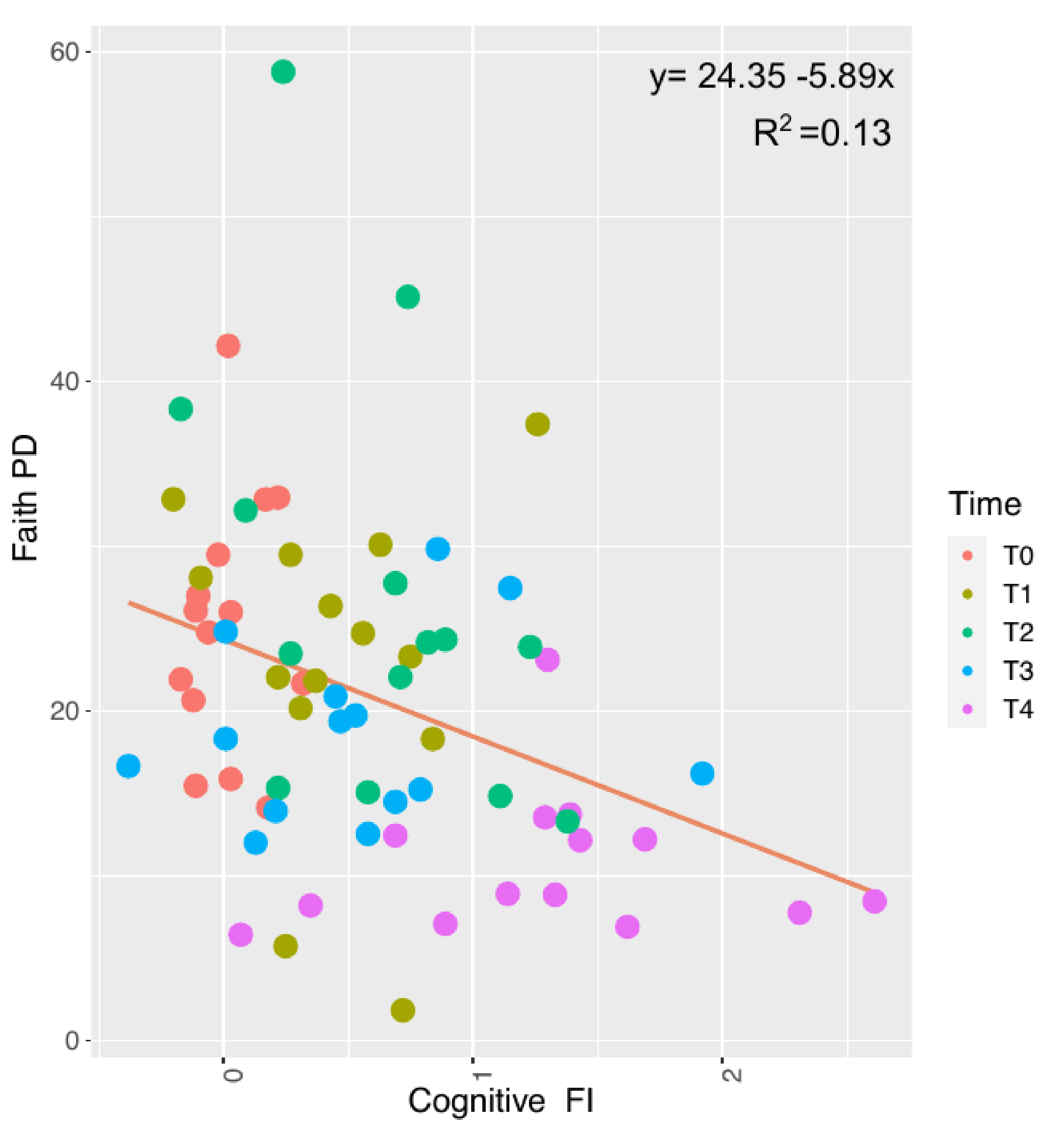
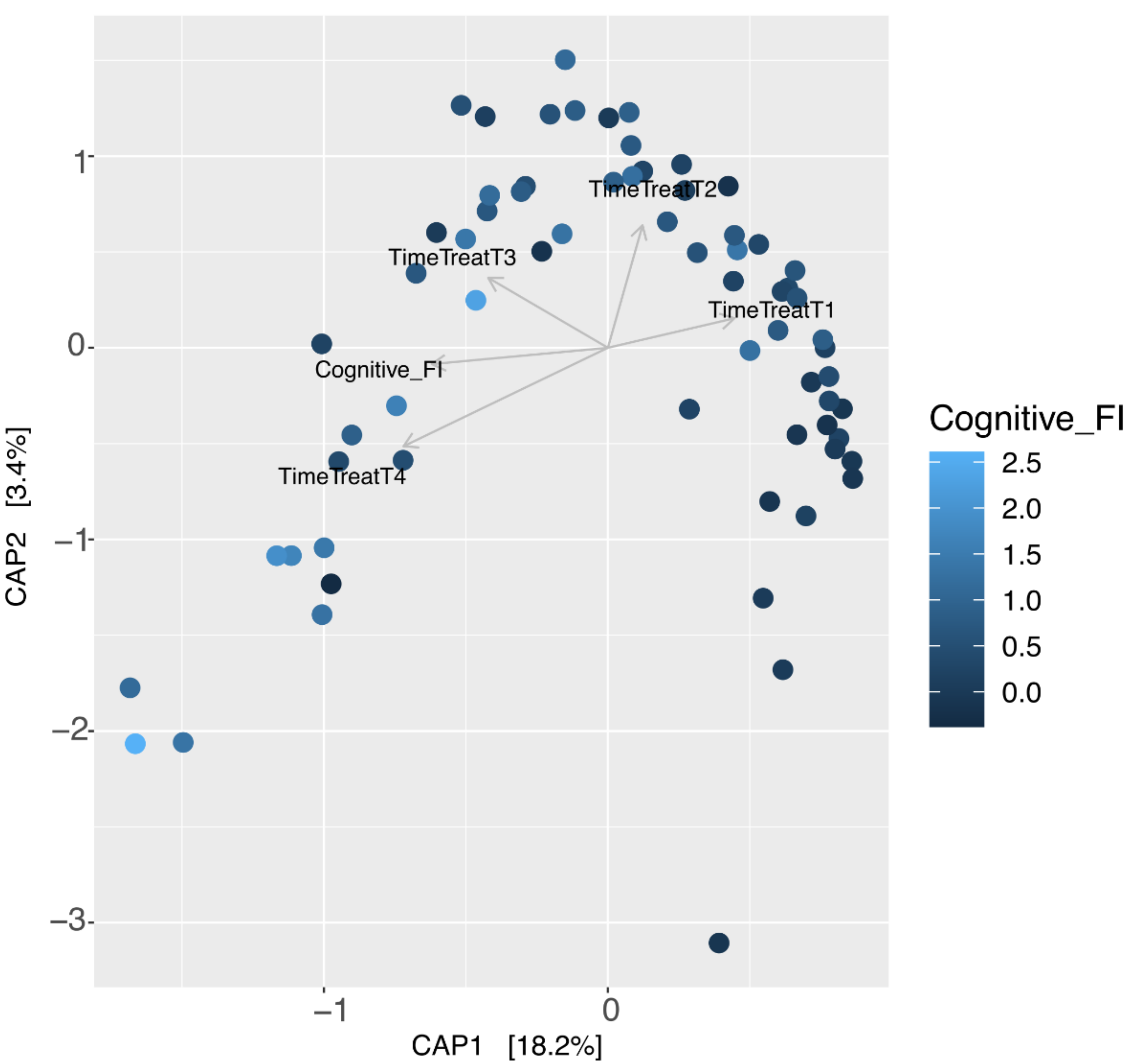
3.4. Aging and Cognitive Frailty Index on Microbiome Composition
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bana, B.; Cabreiro, F. The Microbiome and Aging. Annu. Rev. Genet. 2019, 53, 239–261. [Google Scholar] [CrossRef] [PubMed]
- Ratto, D.; Corana, F.; Mannucci, B.; Priori, E.C.; Cobelli, F.; Roda, E.; Ferrari, B.; Occhinegro, A.; Di Iorio, C.; De Luca, F.; et al. Hericium Erinaceus Improves Recognition Memory and Induces Hippocampal and Cerebellar Neurogenesis in Frail Mice during Aging. Nutrients 2019, 11, 715. [Google Scholar] [CrossRef] [Green Version]
- Burke, S.N.; Ryan, L.; Barnes, C.A. Characterizing Cognitive Aging of Recognition Memory and Related Processes in Animal Models and in Humans. Front. Aging Neurosci. 2012, 4, 15. [Google Scholar] [CrossRef] [Green Version]
- Turriziani, P.; Serra, L.; Fadda, L.; Caltagirone, C.; Carlesimo, G.A. Recollection and Familiarity in Hippocampal Amnesia. Hippocampus 2008, 18, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Rossi, P.; Cesaroni, V.; Brandalise, F.; Occhinegro, A.; Ratto, D.; Perrucci, F.; Lanaia, V.; Girometta, C.; Orrù, G.; Savino, E. Dietary Supplementation of Lion’s Mane Medicinal Mushroom, Hericium erinaceus (Agaricomycetes), and Spatial Memory in Wild-Type Mice. Int. J. Med. Mushrooms 2018, 20, 485–494. [Google Scholar] [CrossRef]
- Cesari, M.; Calvani, R.; Marzetti, E. Frailty in Older Persons. Clin. Geriatr. Med. 2017, 33, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Fried, L.P.; Tangen, C.M.; Walston, J.; Newman, A.B.; Hirsch, C.; Gottdiener, J.; Seeman, T.; Tracy, R.; Kop, W.J.; Burke, G.; et al. Frailty in Older Adults: Evidence for a Phenotype. J. Gerontol. A Biol. Sci. Med. Sci. 2001, 56, M146–M156. [Google Scholar] [CrossRef]
- Bandeen-Roche, K.; Xue, Q.-L.; Ferrucci, L.; Walston, J.; Guralnik, J.M.; Chaves, P.; Zeger, S.L.; Fried, L.P. Phenotype of Frailty: Characterization in the Women’s Health and Aging Studies. J. Gerontol. A Biol. Sci. Med. Sci. 2006, 61, 262–266. [Google Scholar] [CrossRef] [Green Version]
- Piggott, D.A.; Tuddenham, S. The Gut Microbiome and Frailty. Transl. Res. 2020, 221, 23–43. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Cabrera, M.C.; Garcia-Valles, R.; Rodriguez-Mañas, L.; Garcia-Garcia, F.J.; Olaso-Gonzalez, G.; Salvador-Pascual, A.; Tarazona-Santabalbina, F.J.; Viña, J. A New Frailty Score for Experimental Animals Based on the Clinical Phenotype: Inactivity as a Model of Frailty. J. Gerontol. A Biol. Sci. Med. Sci. 2017, 72, 885–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arai, H.; Satake, S.; Kozaki, K. Cognitive Frailty in Geriatrics. Clin. Geriatr. Med. 2018, 34, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Segura, A.; Nehme, J.; Demaria, M. Hallmarks of Cellular Senescence. Trends Cell. Biol. 2018, 28, 436–453. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Wang, Y.; Yuan, M.; Huang, Z.; Zou, Q.; Pu, Y.; Zhao, B.; Cai, Z. Role of Mismatch Repair in Aging. Int. J. Biol. Sci. 2021, 17, 3923–3935. [Google Scholar] [CrossRef]
- Nagpal, R.; Mainali, R.; Ahmadi, S.; Wang, S.; Singh, R.; Kavanagh, K.; Kitzman, D.W.; Kushugulova, A.; Marotta, F.; Yadav, H. Gut Microbiome and Aging: Physiological and Mechanistic Insights. Nutr. Healthy Aging 2018, 4, 267–285. [Google Scholar] [CrossRef] [Green Version]
- DeJong, E.N.; Surette, M.G.; Bowdish, D.M.E. The Gut Microbiota and Unhealthy Aging: Disentangling Cause from Consequence. Cell Host Microbe 2020, 28, 180–189. [Google Scholar] [CrossRef]
- Guarner, F.; Malagelada, J.-R. Gut Flora in Health and Disease. Lancet 2003, 361, 512–519. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the Normal Gut Microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef]
- Shortt, C.; Hasselwander, O.; Meynier, A.; Nauta, A.; Fernández, E.N.; Putz, P.; Rowland, I.; Swann, J.; Türk, J.; Vermeiren, J.; et al. Systematic Review of the Effects of the Intestinal Microbiota on Selected Nutrients and Non-Nutrients. Eur. J. Nutr. 2018, 57, 25–49. [Google Scholar] [CrossRef] [Green Version]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-Bacterial Mutualism in the Human Intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef] [Green Version]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The Human Microbiome Project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Quigley, E.M.M. Gut Bacteria in Health and Disease. Gastroenterol. Hepatol. 2013, 9, 560–569. [Google Scholar]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Rosa, F.; Clerici, M.; Ratto, D.; Occhinegro, A.; Licito, A.; Romeo, M.; Iorio, C.D.; Rossi, P. The Gut-Brain Axis in Alzheimer’s Disease and Omega-3. A Critical Overview of Clinical Trials. Nutrients 2018, 10, 1267. [Google Scholar] [CrossRef] [Green Version]
- Marizzoni, M.; Provasi, S.; Cattaneo, A.; Frisoni, G.B. Microbiota and Neurodegenerative Diseases. Curr. Opin. Neurol. 2017, 30, 630–638. [Google Scholar] [CrossRef]
- Ticinesi, A.; Tana, C.; Nouvenne, A.; Prati, B.; Lauretani, F.; Meschi, T. Gut Microbiota, Cognitive Frailty and Dementia in Older Individuals: A Systematic Review. Clin. Interv. Aging 2018, 13, 1497–1511. [Google Scholar] [CrossRef] [Green Version]
- Brandalise, F.; Cesaroni, V.; Gregori, A.; Repetti, M.; Romano, C.; Orrù, G.; Botta, L.; Girometta, C.; Guglielminetti, M.L.; Savino, E.; et al. Dietary Supplementation of Hericium Erinaceus Increases Mossy Fiber-CA3 Hippocampal Neurotransmission and Recognition Memory in Wild-Type Mice. Evid. Based Complement. Alternat. Med. 2017, 2017, 3864340. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faith, D.P. Science and Philosophy for Molecular Systematics: Which Is the Cart and Which Is the Horse? Mol. Phylogenet. Evol. 2006, 38, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Cuzick, J. A Wilcoxon-Type Test for Trend. Stat. Med. 1985, 4, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Vegan: Community Ecology Package. R Package Vegan, Vers. 2.2-1. Available online: https://www.worldagroforestry.org/publication/vegan-community-ecology-package-r-package-vegan-vers-22-1 (accessed on 5 April 2022).
- Anderson, M.J.; Willis, T.J. Canonical Analysis of Principal Coordinates: A Useful Method of Constrained Ordination for Ecology. Ecology 2003, 84, 511–525. [Google Scholar] [CrossRef]
- Kruskal, J.B. Multidimensional Scaling by Optimizing Goodness of Fit to a Nonmetric Hypothesis. Psychometrika 1964, 29, 1–27. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Foster, Z.S.L.; Sharpton, T.J.; Grünwald, N.J. Metacoder: An R Package for Visualization and Manipulation of Community Taxonomic Diversity Data. PLoS Comput. Biol. 2017, 13, e1005404. [Google Scholar] [CrossRef] [Green Version]
- Ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics—Ggplot2-Package. Available online: https://ggplot2.tidyverse.org/reference/ggplot2-package.html (accessed on 7 July 2022).
- Fox, J.; Weisberg, S. An R Companion to Applied Regression; SAGE Publications: Thousand Oaks, CA, USA, 2011; ISBN 9781412975148. [Google Scholar]
- O’Toole, P.W.; Jeffery, I.B. Gut Microbiota and Aging. Science 2015, 350, 1214–1215. [Google Scholar] [CrossRef]
- Santoro, A.; Ostan, R.; Candela, M.; Biagi, E.; Brigidi, P.; Capri, M.; Franceschi, C. Gut Microbiota Changes in the Extreme Decades of Human Life: A Focus on Centenarians. Cell Mol. Life Sci. 2018, 75, 129–148. [Google Scholar] [CrossRef] [Green Version]
- Wilmanski, T.; Diener, C.; Rappaport, N.; Patwardhan, S.; Wiedrick, J.; Lapidus, J.; Earls, J.C.; Zimmer, A.; Glusman, G.; Robinson, M.; et al. Gut Microbiome Pattern Reflects Healthy Ageing and Predicts Survival in Humans. Nat. Metab. 2021, 3, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Belzer, C.; de Vos, W.M. Akkermansia muciniphila and Its Role in Regulating Host Functions. Microb. Pathog. 2017, 106, 171–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naito, Y.; Uchiyama, K.; Takagi, T. A Next-Generation Beneficial Microbe: Akkermansia muciniphila. J. Clin. Biochem. Nutr. 2018, 63, 33–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macchione, I.G.; Lopetuso, L.R.; Ianiro, G.; Napoli, M.; Gibiino, G.; Rizzatti, G.; Petito, V.; Gasbarrini, A.; Scaldaferri, F. Akkermansia muciniphila: Key Player in Metabolic and Gastrointestinal Disorders. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8075–8083. [Google Scholar] [CrossRef]
- Gerhardt, S.; Mohajeri, M.H. Changes of Colonic Bacterial Composition in Parkinson’s Disease and Other Neurodegenerative Diseases. Nutrients 2018, 10, 708. [Google Scholar] [CrossRef] [Green Version]
- Heintz-Buschart, A.; Pandey, U.; Wicke, T.; Sixel-Döring, F.; Janzen, A.; Sittig-Wiegand, E.; Trenkwalder, C.; Oertel, W.H.; Mollenhauer, B.; Wilmes, P. The Nasal and Gut Microbiome in Parkinson’s Disease and Idiopathic Rapid Eye Movement Sleep Behavior Disorder. Mov. Disord. 2018, 33, 88–98. [Google Scholar] [CrossRef] [Green Version]
- Nishiwaki, H.; Ito, M.; Ishida, T.; Hamaguchi, T.; Maeda, T.; Kashihara, K.; Tsuboi, Y.; Ueyama, J.; Shimamura, T.; Mori, H.; et al. Meta-Analysis of Gut Dysbiosis in Parkinson’s Disease. Mov. Disord. 2020, 35, 1626–1635. [Google Scholar] [CrossRef]
- Ou, Z.; Deng, L.; Lu, Z.; Wu, F.; Liu, W.; Huang, D.; Peng, Y. Protective Effects of Akkermansia muciniphila on Cognitive Deficits and Amyloid Pathology in a Mouse Model of Alzheimer’s Disease. Nutr. Diabetes 2020, 10, 12. [Google Scholar] [CrossRef] [Green Version]
- Higarza, S.G.; Arboleya, S.; Arias, J.L.; Gueimonde, M.; Arias, N. Akkermansia muciniphila and Environmental Enrichment Reverse Cognitive Impairment Associated with High-Fat High-Cholesterol Consumption in Rats. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef]
- Sibai, M.; Altuntaş, E.; Yıldırım, B.; Öztürk, G.; Yıldırım, S.; Demircan, T. Microbiome and Longevity: High Abundance of Longevity-Linked Muribaculaceae in the Gut of the Long-Living Rodent Spalax Leucodon. OMICS 2020, 24, 592–601. [Google Scholar] [CrossRef]
- Smith, B.J.; Miller, R.A.; Ericsson, A.C.; Harrison, D.C.; Strong, R.; Schmidt, T.M. Changes in the Gut Microbiome and Fermentation Products Concurrent with Enhanced Longevity in Acarbose-Treated Mice. BMC Microbiol. 2019, 19, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, F.C.; Wasmund, K.; Cobankovic, I.; Jehmlich, N.; Herbold, C.W.; Lee, K.S.; Sziranyi, B.; Vesely, C.; Decker, T.; Stocker, R.; et al. Rational Design of a Microbial Consortium of Mucosal Sugar Utilizers Reduces Clostridiodes Difficile Colonization. Nat. Commun. 2020, 11, 5104. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Li, Z.; Liu, T.; Yang, N.; Li, Y.; He, J.; Qian, M.; Kuang, Z.; Zhang, W.; Ni, C.; et al. Prebiotics Regulation of Intestinal Microbiota Attenuates Cognitive Dysfunction Induced by Surgery Stimulation in APP/PS1 Mice. Aging Dis. 2020, 11, 1029–1045. [Google Scholar] [CrossRef] [PubMed]
- Shenghua, P.; Ziqin, Z.; Shuyu, T.; Huixia, Z.; Xianglu, R.; Jiao, G. An Integrated Fecal Microbiome and Metabolome in the Aged Mice Reveal Anti-Aging Effects from the Intestines and Biochemical Mechanism of FuFang Zhenshu TiaoZhi(FTZ). Biomed. Pharmacother. 2020, 121, 109421. [Google Scholar] [CrossRef]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The Genus Alistipes: Gut Bacteria With Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Jobin, C. Novel Insights into Microbiome in Colitis and Colorectal Cancer. Curr. Opin. Gastroenterol. 2017, 33, 422–427. [Google Scholar] [CrossRef]
- Naseribafrouei, A.; Hestad, K.; Avershina, E.; Sekelja, M.; Linløkken, A.; Wilson, R.; Rudi, K. Correlation between the Human Fecal Microbiota and Depression. Neurogastroenterol. Motil. 2014, 26, 1155–1162. [Google Scholar] [CrossRef]
- Juckel, G.; Manitz, M.-P.; Freund, N.; Gatermann, S. Impact of Poly I:C Induced Maternal Immune Activation on Offspring’s Gut Microbiome Diversity—Implications for Schizophrenia. Prog. Neuropsychopharmacol. Biol. Psychiatry 2021, 110, 110306. [Google Scholar] [CrossRef]
- Ezeji, J.C.; Sarikonda, D.K.; Hopperton, A.; Erkkila, H.L.; Cohen, D.E.; Martinez, S.P.; Cominelli, F.; Kuwahara, T.; Dichosa, A.E.K.; Good, C.E.; et al. Parabacteroides Distasonis: Intriguing Aerotolerant Gut Anaerobe with Emerging Antimicrobial Resistance and Pathogenic and Probiotic Roles in Human Health. Gut Microbes 2021, 13, 1922241. [Google Scholar] [CrossRef]
- Wang, K.; Liao, M.; Zhou, N.; Bao, L.; Ma, K.; Zheng, Z.; Wang, Y.; Liu, C.; Wang, W.; Wang, J.; et al. Parabacteroides Distasonis Alleviates Obesity and Metabolic Dysfunctions via Production of Succinate and Secondary Bile Acids. Cell Rep. 2019, 26, 222–235.e5. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.-J.; Lin, T.-L.; Tsai, Y.-L.; Wu, T.-R.; Lai, W.-F.; Lu, C.-C.; Lai, H.-C. Next Generation Probiotics in Disease Amelioration. J. Food Drug Anal. 2019, 27, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Nan, X.; Zhao, Y.; Jiang, L.; Wang, H.; Zhang, F.; Hua, D.; Liu, J.; Yao, J.; Yang, L.; et al. Dietary Supplementation of Inulin Ameliorates Subclinical Mastitis via Regulation of Rumen Microbial Community and Metabolites in Dairy Cows. Microbiol. Spectr. 2021, 9, e0010521. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, M.; Yang, M.; Jin, C.; Song, Y.; Chen, J.; Gao, M.; Ai, Z.; Su, D. Pulsatilla Chinensis Saponins Ameliorate Inflammation and DSS-Induced Ulcerative Colitis in Rats by Regulating the Composition and Diversity of Intestinal Flora. Front. Cell Infect. Microbiol. 2021, 11, 728929. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.-D.; Li, Y.; Sun, M.; Yu, C.-J.; Li, J.-Y.; Wang, S.-H.; Yang, D.; Guo, C.-L.; Du, X.; Zhang, W.-J.; et al. Effect of Berberine on Hyperglycaemia and Gut Microbiota Composition in Type 2 Diabetic Goto-Kakizaki Rats. World J. Gastroenterol. 2021, 27, 708–724. [Google Scholar] [CrossRef]
- Lee, D.-Y.; Shin, Y.-J.; Kim, J.-K.; Jang, H.-M.; Joo, M.-K.; Kim, D.-H. Alleviation of Cognitive Impairment by Gut Microbiota Lipopolysaccharide Production-Suppressing Lactobacillus plantarum and Bifidobacterium longum in Mice. Food Funct. 2021, 12, 10750–10763. [Google Scholar] [CrossRef]
- Barandouzi, Z.A.; Starkweather, A.R.; Henderson, W.A.; Gyamfi, A.; Cong, X.S. Altered Composition of Gut Microbiota in Depression: A Systematic Review. Front. Psychiatry 2020, 11, 541. [Google Scholar] [CrossRef]
- Thingholm, L.B.; Rühlemann, M.C.; Koch, M.; Fuqua, B.; Laucke, G.; Boehm, R.; Bang, C.; Franzosa, E.A.; Hübenthal, M.; Rahnavard, A.; et al. Obese Individuals with and without Type 2 Diabetes Show Different Gut Microbial Functional Capacity and Composition. Cell Host Microbe 2019, 26, 252–264.e10. [Google Scholar] [CrossRef]
- Tran, T.T.T.; Cousin, F.J.; Lynch, D.B.; Menon, R.; Brulc, J.; Brown, J.R.-M.; O’Herlihy, E.; Butto, L.F.; Power, K.; Jeffery, I.B.; et al. Prebiotic Supplementation in Frail Older People Affects Specific Gut Microbiota Taxa but Not Global Diversity. Microbiome 2019, 7, 39. [Google Scholar] [CrossRef]
- Wei, J.; Zhao, Y.; Zhou, C.; Zhao, Q.; Zhong, H.; Zhu, X.; Fu, T.; Pan, L.; Shang, Q.; Yu, G. Dietary Polysaccharide from Enteromorpha Clathrata Attenuates Obesity and Increases the Intestinal Abundance of Butyrate-Producing Bacterium, Eubacterium xylanophilum, in Mice Fed a High-Fat Diet. Polymers 2021, 13, 3286. [Google Scholar] [CrossRef]
- Li, K.; Zhang, L.; Xue, J.; Yang, X.; Dong, X.; Sha, L.; Lei, H.; Zhang, X.; Zhu, L.; Wang, Z.; et al. Dietary Inulin Alleviates Diverse Stages of Type 2 Diabetes Mellitus via Anti-Inflammation and Modulating Gut Microbiota in Db/Db Mice. Food Funct. 2019, 10, 1915–1927. [Google Scholar] [CrossRef]
- Shintouo, C.M.; Mets, T.; Beckwee, D.; Bautmans, I.; Ghogomu, S.M.; Souopgui, J.; Leemans, L.; Meriki, H.D.; Njemini, R. Is Inflammageing Influenced by the Microbiota in the Aged Gut? A Systematic Review. Exp. Gerontol. 2020, 141, 111079. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Z.; Ghoshdastidar, S.; Rekha, K.R.; Suresh, D.; Mao, J.; Bivens, N.; Kannan, R.; Joshi, T.; Rosenfeld, C.S.; Upendran, A. Developmental Exposure to Silver Nanoparticles Leads to Long Term Gut Dysbiosis and Neurobehavioral Alterations. Sci. Rep. 2021, 11, 6558. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Li, L.; Schwabacher, A.W.; Young, J.W.; Beitz, D.C. Mechanism of Cholesterol Reduction to Coprostanol by Eubacterium Coprostanoligenes ATCC 51222. Steroids 1996, 61, 33–40. [Google Scholar] [CrossRef]
- Liu, R.T.; Rowan-Nash, A.D.; Sheehan, A.E.; Walsh, R.F.L.; Sanzari, C.M.; Korry, B.J.; Belenky, P. Reductions in Anti-Inflammatory Gut Bacteria Are Associated with Depression in a Sample of Young Adults. Brain Behav. Immun. 2020, 88, 308–324. [Google Scholar] [CrossRef]
- Bai, D.; Sun, T.; Zhao, J.; Du, J.; Bu, X.; Cao, W.; Zhao, Y.; Lu, N. Oroxylin A Maintains the Colonic Mucus Barrier to Reduce Disease Susceptibility by Reconstituting a Dietary Fiber-Deprived Gut Microbiota. Cancer Lett. 2021, 515, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Arias-Jayo, N.; Abecia, L.; Alonso-Sáez, L.; Ramirez-Garcia, A.; Rodriguez, A.; Pardo, M.A. High-Fat Diet Consumption Induces Microbiota Dysbiosis and Intestinal Inflammation in Zebrafish. Microb. Ecol. 2018, 76, 1089–1101. [Google Scholar] [CrossRef]
- Jiang, H.; Ling, Z.; Zhang, Y.; Mao, H.; Ma, Z.; Yin, Y.; Wang, W.; Tang, W.; Tan, Z.; Shi, J.; et al. Altered Fecal Microbiota Composition in Patients with Major Depressive Disorder. Brain Behav. Immun. 2015, 48, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Yun, Y.; An, H.; Zhao, W.; Ma, T.; Wang, Z.; Yang, F. Gut Microbiome and Daytime Function in Chinese Patients with Major Depressive Disorder. J. Psychosom. Res. 2022, 157, 110787. [Google Scholar] [CrossRef]
- Mikami, A.; Ogita, T.; Namai, F.; Shigemori, S.; Sato, T.; Shimosato, T. Oral Administration of Flavonifractor Plautii, a Bacteria Increased With Green Tea Consumption, Promotes Recovery From Acute Colitis in Mice via Suppression of IL-17. Front. Nutr. 2020, 7, 610946. [Google Scholar] [CrossRef]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Blautia-a New Functional Genus with Potential Probiotic Properties? Gut Microbes 2021, 13, 1–21. [Google Scholar] [CrossRef]
- Verhaar, B.J.H.; Hendriksen, H.M.A.; de Leeuw, F.A.; Doorduijn, A.S.; van Leeuwenstijn, M.; Teunissen, C.E.; Barkhof, F.; Scheltens, P.; Kraaij, R.; van Duijn, C.M.; et al. Gut Microbiota Composition Is Related to AD Pathology. Front. Immunol. 2021, 12, 794519. [Google Scholar] [CrossRef]
- Yutin, N.; Galperin, M.Y. A Genomic Update on Clostridial Phylogeny: Gram-Negative Spore Formers and Other Misplaced Clostridia. Environ. Microbiol. 2013, 15, 2631–2641. [Google Scholar] [CrossRef] [PubMed]
- Dandachi, I.; Anani, H.; Hadjadj, L.; Brahimi, S.; Lagier, J.-C.; Daoud, Z.; Rolain, J.-M. Genome Analysis of Lachnoclostridium Phocaeense Isolated from a Patient after Kidney Transplantation in Marseille. New Microbes New Infect. 2021, 41, 100863. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-H.; Wang, M.-F.; Chang, C.-C.; Huang, S.-Y.; Pan, C.-H.; Yeh, Y.-T.; Huang, C.-H.; Chan, C.-H.; Huang, H.-Y. Lacticaseibacillus Paracasei PS23 Effectively Modulates Gut Microbiota Composition and Improves Gastrointestinal Function in Aged SAMP8 Mice. Nutrients 2021, 13, 1116. [Google Scholar] [CrossRef] [PubMed]
- Jena, P.K.; Sheng, L.; Nguyen, M.; Di Lucente, J.; Hu, Y.; Li, Y.; Maezawa, I.; Jin, L.-W.; Wan, Y.-J.Y. Dysregulated Bile Acid Receptor-Mediated Signaling and IL-17A Induction Are Implicated in Diet-Associated Hepatic Health and Cognitive Function. Biomark Res. 2020, 8, 59. [Google Scholar] [CrossRef]
- Bäuerl, C.; Collado, M.C.; Diaz Cuevas, A.; Viña, J.; Pérez Martínez, G. Shifts in Gut Microbiota Composition in an APP/PSS1 Transgenic Mouse Model of Alzheimer’s Disease during Lifespan. Lett. Appl. Microbiol. 2018, 66, 464–471. [Google Scholar] [CrossRef]
- Lawson, P.A.; Citron, D.M.; Tyrrell, K.L.; Finegold, S.M. Reclassification of Clostridium Difficile as Clostridioides Difficile (Hall and O’Toole 1935) Prévot 1938. Anaerobe 2016, 40, 95–99. [Google Scholar] [CrossRef]
- Dürre, P. Physiology and Sporulation in Clostridium. Microbiol. Spectr. 2014, 2, TBS-0010-2012. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Yang, W.-R.; Wang, X.-H.; Li, G.-Q.; Xu, L.-Q.; Cui, X.; Liu, Y.; Zuo, X.-L. Clostridium Butyricum Alleviates Intestinal Low-Grade Inflammation in TNBS-Induced Irritable Bowel Syndrome in Mice by Regulating Functional Status of Lamina Propria Dendritic Cells. World J. Gastroenterol. 2019, 25, 5469–5482. [Google Scholar] [CrossRef]
- Kashiwagi, I.; Morita, R.; Schichita, T.; Komai, K.; Saeki, K.; Matsumoto, M.; Takeda, K.; Nomura, M.; Hayashi, A.; Kanai, T.; et al. Smad2 and Smad3 Inversely Regulate TGF-β Autoinduction in Clostridium Butyricum-Activated Dendritic Cells. Immunity 2015, 43, 65–79. [Google Scholar] [CrossRef] [Green Version]
- Rom, O.; Liu, Y.; Liu, Z.; Zhao, Y.; Wu, J.; Ghrayeb, A.; Villacorta, L.; Fan, Y.; Chang, L.; Wang, L.; et al. Glycine-Based Treatment Ameliorates NAFLD by Modulating Fatty Acid Oxidation, Glutathione Synthesis, and the Gut Microbiome. Sci. Transl. Med. 2020, 12, eaaz2841. [Google Scholar] [CrossRef]
- Zeng, Q.; Li, D.; He, Y.; Li, Y.; Yang, Z.; Zhao, X.; Liu, Y.; Wang, Y.; Sun, J.; Feng, X.; et al. Discrepant Gut Microbiota Markers for the Classification of Obesity-Related Metabolic Abnormalities. Sci. Rep. 2019, 9, 13424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco-de-Moraes, A.C.; de Almeida-Pititto, B.; da Rocha Fernandes, G.; Gomes, E.P.; da Costa Pereira, A.; Ferreira, S.R.G. Worse Inflammatory Profile in Omnivores than in Vegetarians Associates with the Gut Microbiota Composition. Diabetol. Metab. Syndr. 2017, 9, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ialenti, A.; Di Meglio, P.; Grassia, G.; Maffia, P.; Di Rosa, M.; Lanzetta, R.; Molinaro, A.; Silipo, A.; Grant, W.; Ianaro, A. A Novel Lipid A from Halomonas Magadiensis Inhibits Enteric LPS-Induced Human Monocyte Activation. Eur. J. Immunol. 2006, 36, 354–360. [Google Scholar] [CrossRef]
- Van Soest, A.P.M.; Hermes, G.D.A.; Berendsen, A.A.M.; van de Rest, O.; Zoetendal, E.G.; Fuentes, S.; Santoro, A.; Franceschi, C.; de Groot, L.C.P.G.M.; de Vos, W.M. Associations between Pro- and Anti-Inflammatory Gastro-Intestinal Microbiota, Diet, and Cognitive Functioning in Dutch Healthy Older Adults: The NU-AGE Study. Nutrients 2020, 12, 3471. [Google Scholar] [CrossRef]
- Henke, M.T.; Kenny, D.J.; Cassilly, C.D.; Vlamakis, H.; Xavier, R.J.; Clardy, J. Ruminococcus Gnavus, a Member of the Human Gut Microbiome Associated with Crohn’s Disease, Produces an Inflammatory Polysaccharide. Proc. Natl. Acad. Sci. USA 2019, 116, 12672–12677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beaud, D.; Tailliez, P.; Anba-Mondoloni, J. Genetic Characterization of the Beta-Glucuronidase Enzyme from a Human Intestinal Bacterium, Ruminococcus Gnavus. Microbiology 2005, 151, 2323–2330. [Google Scholar] [CrossRef]
- Chahwan, B.; Kwan, S.; Isik, A.; van Hemert, S.; Burke, C.; Roberts, L. Gut Feelings: A Randomised, Triple-Blind, Placebo-Controlled Trial of Probiotics for Depressive Symptoms. J. Affect. Disord. 2019, 253, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Ponziani, F.R.; Calvani, R.; Marini, F.; Biancolillo, A.; Coelho-Junior, H.J.; Gervasoni, J.; Primiano, A.; Putignani, L.; Del Chierico, F.; et al. Gut Microbial, Inflammatory and Metabolic Signatures in Older People with Physical Frailty and Sarcopenia: Results from the BIOSPHERE Study. Nutrients 2019, 12, 65. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, Z.-Q.; Shen, L.-L.; Li, W.-W.; Fu, X.; Zeng, F.; Gui, L.; Lü, Y.; Cai, M.; Zhu, C.; Tan, Y.-L.; et al. Gut Microbiota Is Altered in Patients with Alzheimer’s Disease. J. Alzheimers Dis. 2018, 63, 1337–1346. [Google Scholar] [CrossRef] [Green Version]
- Ren, T.; Gao, Y.; Qiu, Y.; Jiang, S.; Zhang, Q.; Zhang, J.; Wang, L.; Zhang, Y.; Wang, L.; Nie, K. Gut Microbiota Altered in Mild Cognitive Impairment Compared With Normal Cognition in Sporadic Parkinson’s Disease. Front. Neurol. 2020, 11, 137. [Google Scholar] [CrossRef] [PubMed]
- Khine, W.W.T.; Voong, M.L.; Ng, T.K.S.; Feng, L.; Rane, G.A.; Kumar, A.P.; Kua, E.H.; Mahendran, R.; Mahendran, R.; Lee, Y.-K. Mental Awareness Improved Mild Cognitive Impairment and Modulated Gut Microbiome. Aging 2020, 12, 24371–24393. [Google Scholar] [CrossRef] [PubMed]
- Mudd, A.T.; Berding, K.; Wang, M.; Donovan, S.M.; Dilger, R.N. Serum Cortisol Mediates the Relationship between Fecal Ruminococcus and Brain N-Acetylaspartate in the Young Pig. Gut Microbes 2017, 8, 589–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, R.; Guan, X.; Huang, K.; Zhang, Y.; Li, S.; Xia, J.; Shen, M. Flavonoids from Whole-Grain Oat Alleviated High-Fat Diet-Induced Hyperlipidemia via Regulating Bile Acid Metabolism and Gut Microbiota in Mice. J. Agric. Food Chem. 2021, 69, 7629–7640. [Google Scholar] [CrossRef] [PubMed]
- Kenna, J.E.; Chua, E.G.; Bakeberg, M.; Tay, A.; McGregor, S.; Gorecki, A.; Horne, M.; Marshall, B.; Mastaglia, F.L.; Anderton, R.S. Changes in the Gut Microbiome and Predicted Functional Metabolic Effects in an Australian Parkinson’s Disease Cohort. Front. Neurosci. 2021, 15, 756951. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Mo, X.; Huang, H.; Chen, X.; Liu, H.; Peng, Z.; Chen, L.; Rong, S.; Yang, W.; Xu, S.; et al. Yeast β-Glucan Alleviates Cognitive Deficit by Regulating Gut Microbiota and Metabolites in Aβ1-42-Induced AD-like Mice. Int. J. Biol. Macromol. 2020, 161, 258–270. [Google Scholar] [CrossRef]
- Kong, C.; Gao, R.; Yan, X.; Huang, L.; Qin, H. Probiotics Improve Gut Microbiota Dysbiosis in Obese Mice Fed a High-Fat or High-Sucrose Diet. Nutrition 2019, 60, 175–184. [Google Scholar] [CrossRef]
- El Aidy, S.; Van den Abbeele, P.; Van de Wiele, T.; Louis, P.; Kleerebezem, M. Intestinal Colonization: How Key Microbial Players Become Established in This Dynamic Process: Microbial Metabolic Activities and the Interplay between the Host and Microbes. Bioessays 2013, 35, 913–923. [Google Scholar] [CrossRef]
- Wang, J.; Ji, H.; Wang, S.; Liu, H.; Zhang, W.; Zhang, D.; Wang, Y. Probiotic Lactobacillus plantarum Promotes Intestinal Barrier Function by Strengthening the Epithelium and Modulating Gut Microbiota. Front. Microbiol. 2018, 9, 1953. [Google Scholar] [CrossRef] [Green Version]
- Jeong, S.; Huang, L.-K.; Tsai, M.-J.; Liao, Y.-T.; Lin, Y.-S.; Hu, C.-J.; Hsu, Y.-H. Cognitive Function Associated with Gut Microbial Abundance in Sucrose and S-Adenosyl-L-Methionine (SAMe) Metabolic Pathways. J. Alzheimers Dis. 2022, 87, 1115–1130. [Google Scholar] [CrossRef]
- Qian, Y.; Yang, X.; Xu, S.; Wu, C.; Song, Y.; Qin, N.; Chen, S.-D.; Xiao, Q. Alteration of the Fecal Microbiota in Chinese Patients with Parkinson’s Disease. Brain Behav. Immun. 2018, 70, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Terzo, S.; Mulè, F.; Caldara, G.F.; Baldassano, S.; Puleio, R.; Vitale, M.; Cassata, G.; Ferrantelli, V.; Amato, A. Pistachio Consumption Alleviates Inflammation and Improves Gut Microbiota Composition in Mice Fed a High-Fat Diet. Int. J. Mol. Sci. 2020, 21, 365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Amakye, W.K.; Gong, C.; Ren, Z.; Yuan, E.; Ren, J. Effect of Oral and Intraperitoneal Administration of Walnut-Derived Pentapeptide PW5 on Cognitive Impairments in APPSWE/PS1ΔE9 Mice. Free Radic. Biol. Med. 2022, 180, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, Y.; Deng, Y.; Liang, S.; He, Y.; Chen, Y.; Liu, C.; Lin, C.; Han, L.; Tu, G.; et al. Chaihu-Shugan-San Decoction Modulates Intestinal Microbe Dysbiosis and Alleviates Chronic Metabolic Inflammation in NAFLD Rats via the NLRP3 Inflammasome Pathway. Evid. Based Complement. Alternat. Med. 2018, 2018, 9390786. [Google Scholar] [CrossRef] [Green Version]
- Monk, J.M.; Lepp, D.; Zhang, C.P.; Wu, W.; Zarepoor, L.; Lu, J.T.; Pauls, K.P.; Tsao, R.; Wood, G.A.; Robinson, L.E.; et al. Diets Enriched with Cranberry Beans Alter the Microbiota and Mitigate Colitis Severity and Associated Inflammation. J. Nutr. Biochem. 2016, 28, 129–139. [Google Scholar] [CrossRef]
- Kim, S.; Choi, S.; Dutta, M.; Asubonteng, J.O.; Polunas, M.; Goedken, M.; Gonzalez, F.J.; Cui, J.Y.; Gyamfi, M.A. Pregnane X Receptor Exacerbates Nonalcoholic Fatty Liver Disease Accompanied by Obesity- and Inflammation-Prone Gut Microbiome Signature. Biochem. Pharmacol. 2021, 193, 114698. [Google Scholar] [CrossRef]
- O’Callaghan, J.; O’Toole, P.W. Lactobacillus: Host-Microbe Relationships. Curr. Top Microbiol. Immunol. 2013, 358, 119–154. [Google Scholar] [CrossRef]
- Pessione, E. Lactic Acid Bacteria Contribution to Gut Microbiota Complexity: Lights and Shadows. Front. Cell Infect. Microbiol. 2012, 2, 86. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; d’Aigle, J.; Atadja, L.; Quaicoe, V.; Honarpisheh, P.; Ganesh, B.P.; Hassan, A.; Graf, J.; Petrosino, J.; Putluri, N.; et al. Gut Microbiota-Derived Short-Chain Fatty Acids Promote Poststroke Recovery in Aged Mice. Circ. Res. 2020, 127, 453–465. [Google Scholar] [CrossRef]
- Emge, J.R.; Huynh, K.; Miller, E.N.; Kaur, M.; Reardon, C.; Barrett, K.E.; Gareau, M.G. Modulation of the Microbiota-Gut-Brain Axis by Probiotics in a Murine Model of Inflammatory Bowel Disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 310, G989–G998. [Google Scholar] [CrossRef] [Green Version]
- Van Tongeren, S.P.; Slaets, J.P.J.; Harmsen, H.J.M.; Welling, G.W. Fecal Microbiota Composition and Frailty. Appl. Environ. Microbiol. 2005, 71, 6438–6442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aizawa, E.; Tsuji, H.; Asahara, T.; Takahashi, T.; Teraishi, T.; Yoshida, S.; Ota, M.; Koga, N.; Hattori, K.; Kunugi, H. Possible Association of Bifidobacterium and Lactobacillus in the Gut Microbiota of Patients with Major Depressive Disorder. J. Affect. Disord. 2016, 202, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.-J.; Shen, Y.-E.; Wang, X.; Fu, S.; Zhang, X.; Zhang, Y.-N.; Wang, R.-T. Concomitant Memantine and Lactobacillus Plantarum Treatment Attenuates Cognitive Impairments in APP/PS1 Mice. Aging 2020, 12, 628–649. [Google Scholar] [CrossRef] [PubMed]
- Lew, L.-C.; Hor, Y.-Y.; Yusoff, N.A.A.; Choi, S.-B.; Yusoff, M.S.B.; Roslan, N.S.; Ahmad, A.; Mohammad, J.A.M.; Abdullah, M.F.I.L.; Zakaria, N.; et al. Probiotic Lactobacillus Plantarum P8 Alleviated Stress and Anxiety While Enhancing Memory and Cognition in Stressed Adults: A Randomised, Double-Blind, Placebo-Controlled Study. Clin. Nutr. 2019, 38, 2053–2064. [Google Scholar] [CrossRef]
- Smith, C.J.; Emge, J.R.; Berzins, K.; Lung, L.; Khamishon, R.; Shah, P.; Rodrigues, D.M.; Sousa, A.J.; Reardon, C.; Sherman, P.M.; et al. Probiotics Normalize the Gut-Brain-Microbiota Axis in Immunodeficient Mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, G793–G802. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kim, J.; Park, S.-D.; Shim, J.-J.; Lee, J.-L. Lactobacillus Plantarum HY7715 Ameliorates Sarcopenia by Improving Skeletal Muscle Mass and Function in Aged Balb/c Mice. Int. J. Mol. Sci. 2021, 22, 10023. [Google Scholar] [CrossRef]
- Valcheva, R.; Hotte, N.; Gillevet, P.; Sikaroodi, M.; Thiessen, A.; Madsen, K.L. Soluble Dextrin Fibers Alter the Intestinal Microbiota and Reduce Proinflammatory Cytokine Secretion in Male IL-10-Deficient Mice. J. Nutr. 2015, 145, 2060–2066. [Google Scholar] [CrossRef] [Green Version]
- Long-Smith, C.; O’Riordan, K.J.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Microbiota-Gut-Brain Axis: New Therapeutic Opportunities. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 477–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Shannon Index | ||
|---|---|---|
| Combination | WSRT | FDR p-Value |
| T0–T1 | 17 | 0.092 |
| T1–T2 | 6 | 0.007 |
| T2–T3 | 13 | 0.011 |
| T3–T4 | 41 | 0.787 |
| Faith’s PD Index | ||
| Combination | WSRT | FDR p-Value |
| T0–T1 | 36 | 0.850 |
| T1–T2 | 31 | 0.569 |
| T2–T3 | 24 | 0.0785 |
| T3–T4 | 0 | 0.0002 |
| Combination | SumsOfSqs | MeanSqs | F.Model | R2 | p-Value. Corrected |
| T0–T1 | 0.191 | 0.191 | 2.200 | 0.084 | 0.029 |
| T1–T2 | 0.205 | 0.205 | 2.317 | 0.088 | 0.004 |
| T1–T3 | 0.746 | 0.746 | 6.188 | 0.205 | 0.001 |
| T2–T3 | 0.351 | 0.351 | 2.516 | 0.088 | 0.001 |
| T3–T4 | 0.301 | 0.301 | 1.555 | 0.059 | 0.109 |
| Genera | Published Effects on Host Health | Changes with Cognitive Frailty Increase | Possible Role |
|---|---|---|---|
| Parabacteroides | Dichotomous role demonstrated also in CNS [62,63,64]. | Decrease | Dysbiotic/Adaptive |
| Clostridia UCG-014 | Pro-inflammatory [65,66,67]. | Decrease | Adaptive |
| Oscillibacter | Negatively related to cognitive performance [68]; related to depression [60,69], metabolic disease [70], and inflammation [71]. | Decrease | Adaptive |
| Mucispirillum | Opportunistic pathogens [72]; positively related to inflammation, LPS [72,73,74], and changes in different behavioral domains [75]. | Decrease | Dysbiotic |
| Coprostanoligenes | Negatively related to inflammation [76,77,78]. | Decrease | Dysbiotic |
| Flavonifractor | Dichotomous role, also demonstrated in CNS [79,80,81,82]. | Decrease | Dysbiotic/Adaptive |
| Blautia | Butyrate producer with probiotic potential [83]. | Decrease | Dysbiotic |
| Monoglobus | Negatively related to amyloid presence in the brain [84]. | Decrease | Dysbiotic |
| Lachnoclostridium | Butyrate producer [85,86]. | Decrease | Dysbiotic |
| Oscillospiraceae UCG_005 | Negatively related to inflammation [81]. | Decrease | Dysbiotic |
| Marvinbryantia | Negatively related to amyloid presence in the brain [84]. | Decrease | Dysbiotic |
| Erysipelotrichaceae | Pro-inflammatory [87,88,89]; enriched in mouse AD model and senescence-accelerated mice model [87,89]. | Decrease | Adaptive |
| Clostridium sensu stricto 1, known also as C. butyricum [90] | Dichotomous role [91,92,93,94,95]. | Decrease | Dysbiotic/Adaptive |
| Halomonas | Dichotomous role [96,97]. | Decrease | Dysbiotic/Adaptive |
| Ruminococcus | Pro-inflammatory [98,99,100]; dysregulated in patients with depression, physical frailty and sarcopenia, AD, and PD and MCI [80,101,102,103,104]; risk indicator of MCI [105]; negatively related to cognitive performance [103,104,105] and to marker of neuronal health [106]. | Increase | Dysbiotic |
| Colidextribacter | Positively related to inflammation [65,107]; decreased in PD patients [108]. | Increase | Dysbiotic |
| Anaerotruncus | Dichotomous role [109,110,111,112], also in CNS (AD, PD, and cognitive impairment) [109,113,114]. | Increase | Dysbiotic/Adaptive |
| Anaeroplasma | Healthy bacterium [115] decreased in AD mice model [116] and in obese rats [117]; negatively related to inflammation [118]. | Increase | Adaptive |
| Peptococcaceae | Pro-inflammatory [118,119]. | Increase | Dysbiotic |
| Lactobacillus | Anti-inflammatory, butyrate producer with probiotic effect [120,121,122], its beneficial effects are demonstrated also in CNS (altered behaviors, depression, anxiety, stress, cognitive impairment, andAD models) [123,124,125,126,127,128]; reduced sarcopenic process and physical frailty in mice [129]. | Increase | Adaptive |
| Ruminococcaceae incertae sedis | Anti-inflammatory, butyrate producer [130]. | Increase | Adaptive |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ratto, D.; Roda, E.; Romeo, M.; Venuti, M.T.; Desiderio, A.; Lupo, G.; Capelli, E.; Sandionigi, A.; Rossi, P. The Many Ages of Microbiome–Gut–Brain Axis. Nutrients 2022, 14, 2937. https://doi.org/10.3390/nu14142937
Ratto D, Roda E, Romeo M, Venuti MT, Desiderio A, Lupo G, Capelli E, Sandionigi A, Rossi P. The Many Ages of Microbiome–Gut–Brain Axis. Nutrients. 2022; 14(14):2937. https://doi.org/10.3390/nu14142937
Chicago/Turabian StyleRatto, Daniela, Elisa Roda, Marcello Romeo, Maria Teresa Venuti, Anthea Desiderio, Giuseppe Lupo, Enrica Capelli, Anna Sandionigi, and Paola Rossi. 2022. "The Many Ages of Microbiome–Gut–Brain Axis" Nutrients 14, no. 14: 2937. https://doi.org/10.3390/nu14142937







