Spore Powder of Paecilomyces hepiali Shapes Gut Microbiota to Relieve Exercise-Induced Fatigue in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Mycelium and Spores of Paecilomyces hepiali
2.2. Metabolite Extraction and Liquid Chromatography with Tandem Mass Spectrometry (LC-MS/MS) Analysis
2.3. Animals and Treatment
2.4. Sample Collection and Biomarker Determination
2.5. Histopathological Examination
2.6. Gut Microbiota Analysis by 16S rRNA Gene Sequencing
2.7. Statistical Analysis
3. Results
3.1. Characteristics of Spores and Mycelia of Paecilomyces hepiali
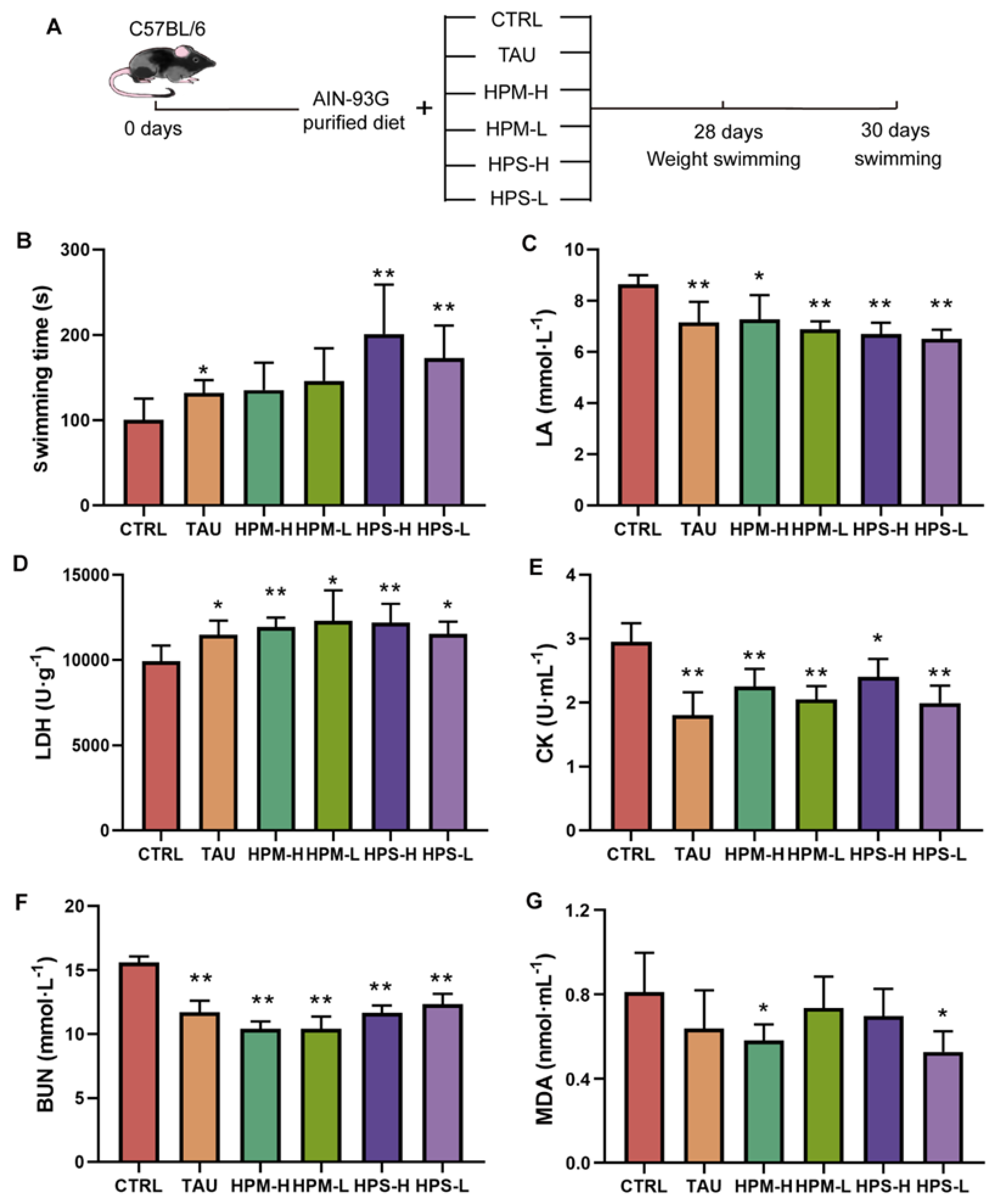
3.2. The Anti-Fatigue Effects of the Spore and Mycelia of Paecilomyces hepiali
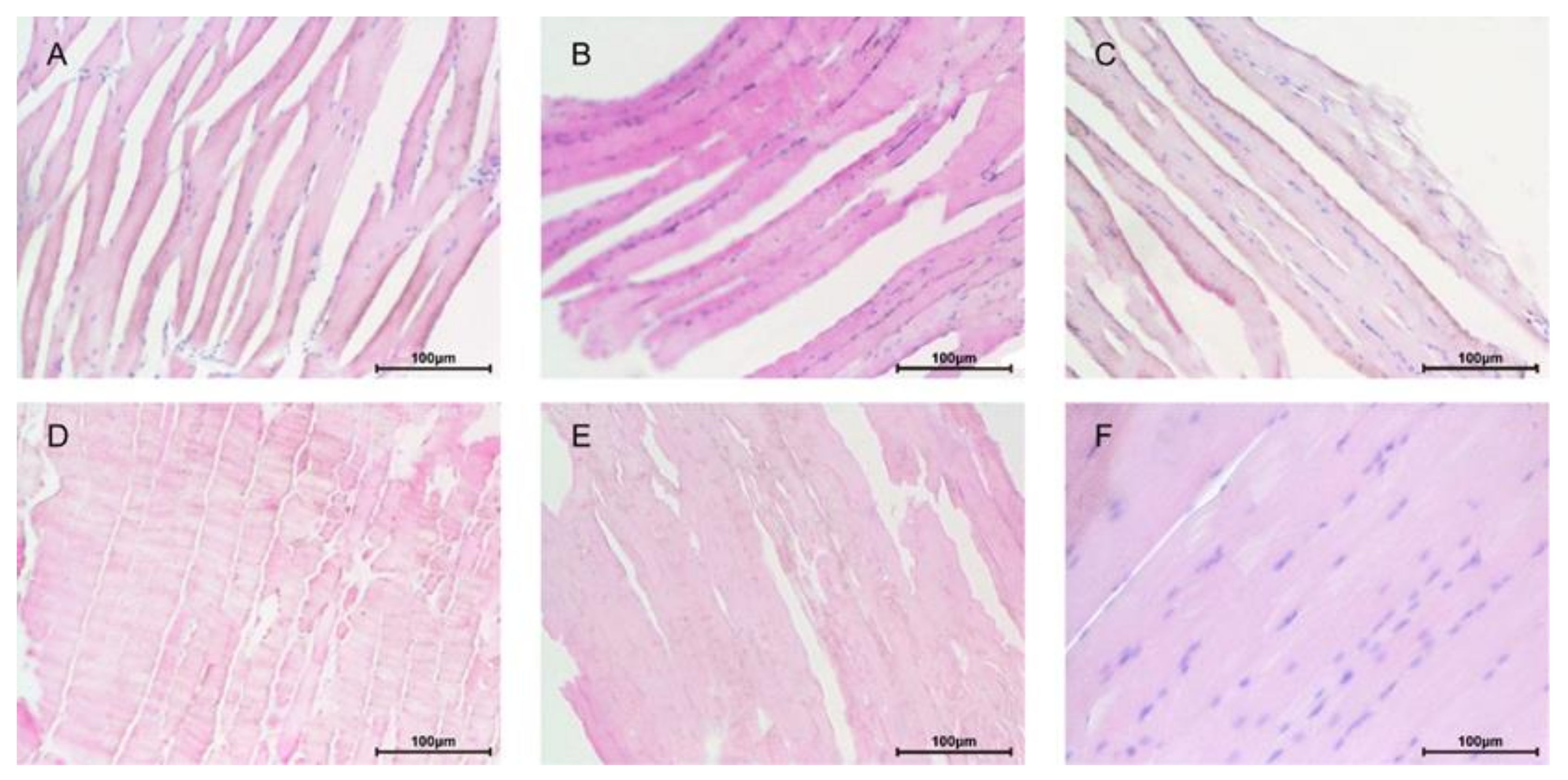
3.3. Effects of the Spore and Mycelia of Paecilomyces hepiali on Blood Biochemical Indicators and Muscle Histopathology
3.4. Effects of the Spore and Mycelia of Paecilomyces hepiali on the Oxidative Stress and Glycogen Content
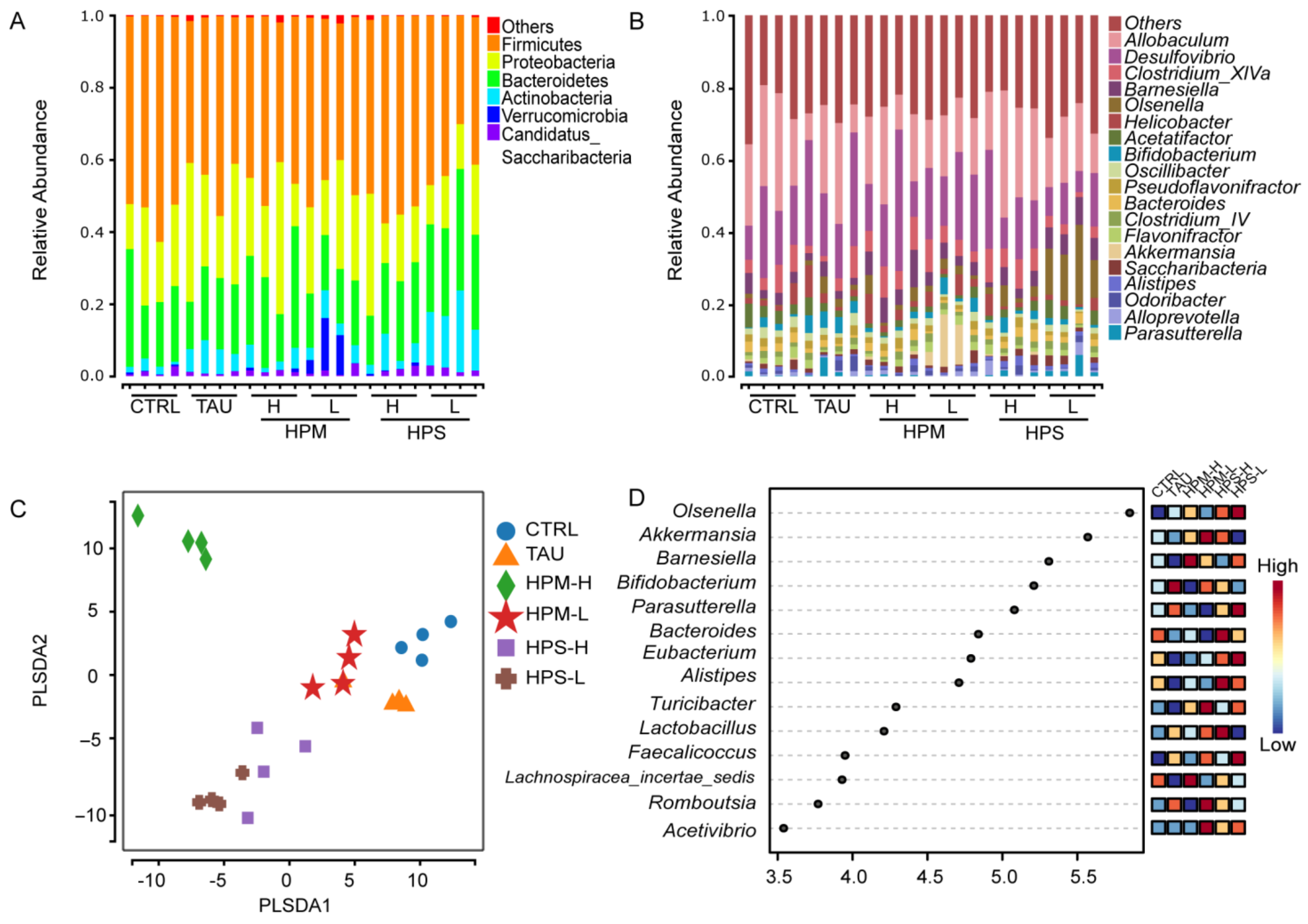
3.5. Effects of the Spore and Mycelia of Paecilomyces hepiali on Gut Microbiota
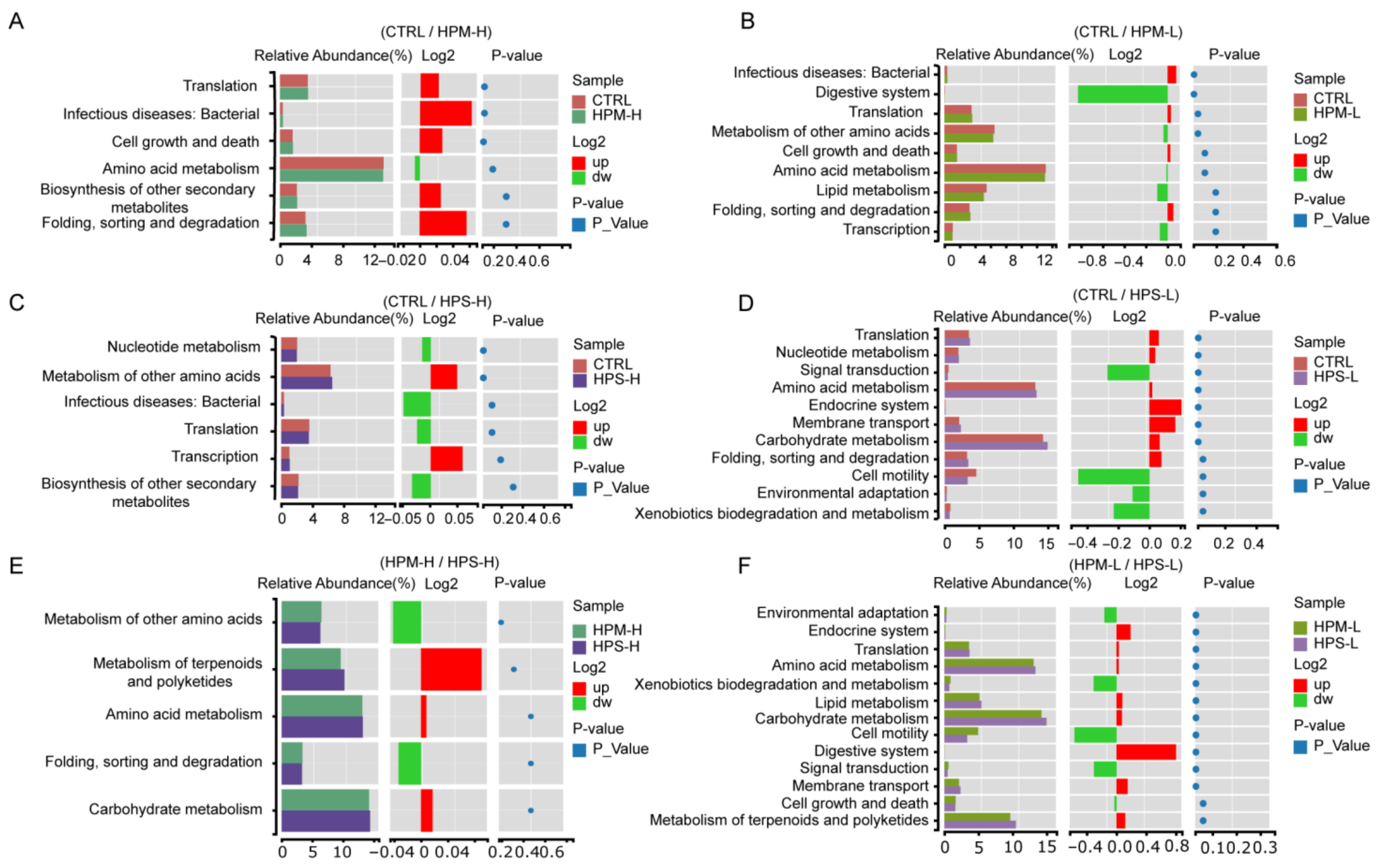
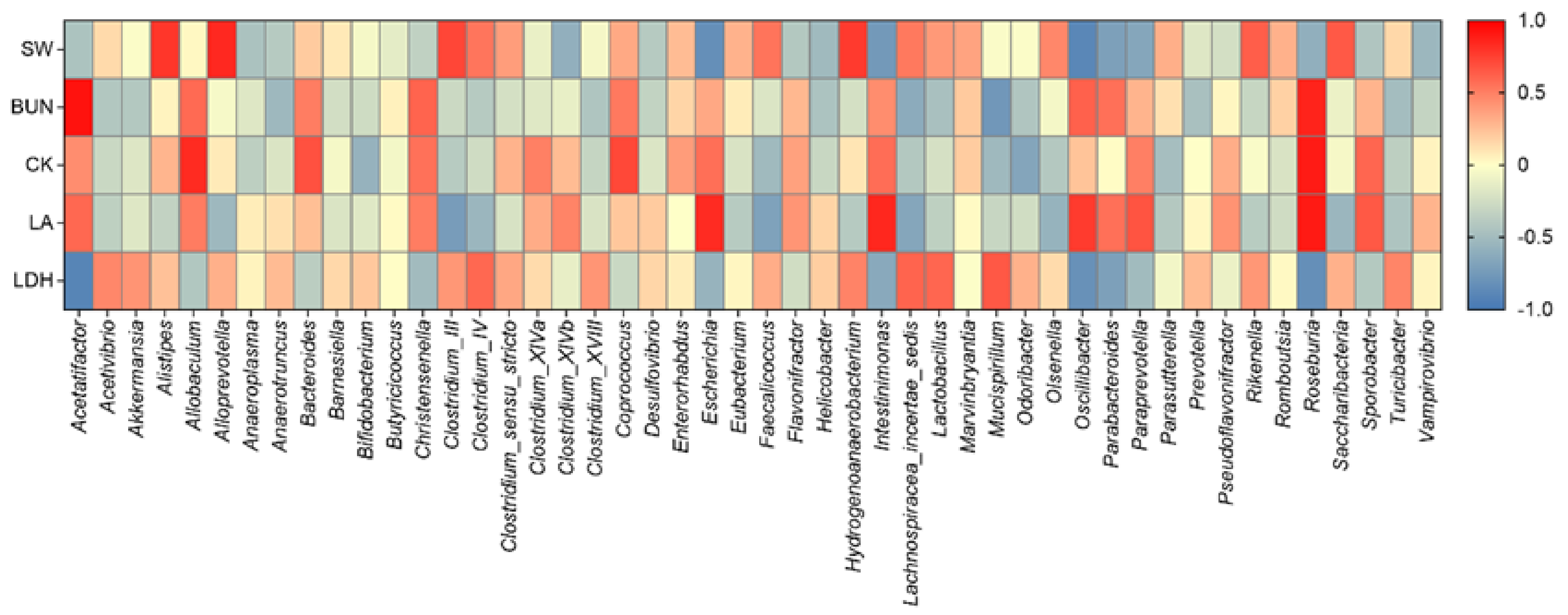
3.6. Correlation between Gut Microbiota and Anti-Fatigue Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiang, X.; Chu, Q.B.; Li, L.Z.; Qin, L.Y.; Hao, J.; Kou, L.; Lin, F.; Wang, D. The anti-fatigue activities of Tuber melanosporum in a mouse model. Exp. Ther. Med. 2018, 15, 3066–3073. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.X.; Zeng, H.L.; Lin, S.L.; Zhang, Z.G.; Zhang, Y.; Hu, J.M. Anti-fatigue activities of hairtail (Trichiurus lepturus) hydrolysate in an endurance swimming mice model. J. Funct. Foods 2020, 74, 104207. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, A.; Yang, X. Effect of lemon seed flavonoids on the anti-fatigue and antioxidant effects of exhausted running exercise mice. J. Food Biochem. 2021, 45, e13620. [Google Scholar] [CrossRef]
- Li, Y.; Deng, Y.; Li, Z.; Liu, Z.; Piao, M.; Cui, X. Composition, physicochemical properties, and anti-fatigue activity of water-soluble okra (Abelmoschus esculentus) stem pectins. Int. J. Biol. Macromol. 2020, 165, 2630–2639. [Google Scholar] [CrossRef]
- Chang, Y.B.; Hong, K.-B.; Kim, M.G.; Suh, H.J.; Jo, K. Effect of the protein hydrolysate of rice syrup meal on the endurance exercise performance of BALB/c mice. Food Funct. 2021, 12, 1338–1348. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-J.; Jhang, W.-L.; Lee, M.-C.; Bat-Otgon, B.; Narantungalag, E.; Huang, C.-C. Lactose-riched Mongolian mare’s milk improves physical fatigue and exercise performance in mice. Int. J. Med. Sci. 2021, 18, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Ren, J.-w.; Zhang, T.; Liu, R.; Wu, L.; Du, Q.; Li, Y. Anti-fatigue effects of small-molecule oligopeptides isolated from Panax quinquefolium L. in mice. Food Funct. 2018, 9, 4266–4273. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Lin, L.; Chen, H.; Ge, X.; Huang, Y.; Zheng, Z.; Li, S.; Pan, Y.; Liu, B.; Zeng, F. Anti-fatigue property of the oyster polypeptide fraction and its effect on gut microbiota in mice. Food Funct. 2020, 11, 8659–8669. [Google Scholar] [CrossRef] [PubMed]
- Nay, K.; Jollet, M.; Goustard, B.; Baati, N.; Vernus, B.; Pontones, M.; Lefeuvre-Orfila, L.; Bendavid, C.; Rue, O.; Mariadassou, M.; et al. Gut bacteria are critical for optimal muscle function: A potential link with glucose homeostasis. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E158–E171. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.J.; Huang, W.C.; Lin, J.S.; Chen, Y.M.; Ho, S.T.; Huang, C.C.; Tung, Y.T. Kefir supplementation modifies gut microbiota composition, reduces physical fatigue, and improves exercise performance in mice. Nutrients 2018, 10, 862. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.; Ma, N.; Zheng, H.; Ma, G.; Zhao, L.; Hu, Q. Tuber indicum polysaccharide relieves fatigue by regulating gut microbiota in mice. J. Funct. Foods 2019, 63, 103580. [Google Scholar] [CrossRef]
- Chen, Y.-M.; Chiu, W.-C.; Chiu, Y.-S.; Li, T.; Sung, H.-C.; Hsiao, C.-Y. Supplementation of nano-bubble curcumin extract improves gut microbiota composition and exercise performance in mice. Food Funct. 2020, 11, 3574–3584. [Google Scholar] [CrossRef]
- Hopping, K.A.; Chignell, S.M.; Lambin, E.F. The demise of caterpillar fungus in the Himalayan region due to climate change and overharvesting. Proc. Natl. Acad. Sci. USA 2018, 115, 11489–11494. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Huang, T.; Chen, C.; Ma, Y. Research Status on Paecilomyces Hepialid. Chin. J. Inf. Tradit. Chin. Med. 2015, 22, 129–131. [Google Scholar]
- Dan, A.K.; Hu, Y.J.; Chen, R.Y.; Lin, X.Y.; Tian, Y.Q.; Wang, S.Y. Advances in research on chemical constituents and pharmacological effects of Paecilomyces hepiali. Food Sci. Hum. Wellness 2021, 10, 401–407. [Google Scholar] [CrossRef]
- Teng, W.-Z.; Song, J.; Meng, F.-X.; Meng, Q.-F.; Lu, J.-H.; Hu, S.; Teng, L.-R.; Wang, D.; Xie, J. Study on the Detection of Active Ingredient Contents of Paecilomyces hepiali Mycelium via Near Infrared Spectroscopy. Spectrosc. Spectr. Anal. 2014, 34, 2645–2651. [Google Scholar] [CrossRef]
- Luo, W.; Yang, H. Determination of Some Chemical Components of Fermented Cordyceps Mycelia Using Near-Infrared Spectroscopy. Philipp. Agric. Sci. 2013, 96, 137–143. [Google Scholar]
- Sayed-Hassan-Raza, S.; Zhenming, L.; Shuoshuo, L.; Huaxiang, L.; Zhishan, L.; Xiaojuan, Z.; Jinsong, S.; Zhenghong, X. Recycling fermentation method of Paecilomyces hepiali based on asexual reproduction. China Brew. 2018, 37, 34–38. [Google Scholar] [CrossRef]
- Wang, J.; Li, L.Z.; Liu, Y.G.; Teng, L.R.; Lu, J.H.; Xie, J.; Hi, W.J.; Liu, Y.; Wang, D.; Teng, L.S. Investigations on the antifatigue and antihypoxic effects of Paecilomyces hepiali extract. Mol. Med. Rep. 2016, 13, 1861–1868. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Liang, H.; Wen, Q.; Ye, E.; Yang, Z. Effect of Paecilomyces hepiali mycelium on physical fatigue and hypoxia in mice. Mil. Med. Sci. 2014, 38, 784–790. [Google Scholar]
- Li, H.-X.; Lu, Z.-M.; Geng, Y.; Gong, J.-S.; Zhang, X.-J.; Shi, J.-S.; Xu, Z.-H.; Ma, Y.-H. Efficient production of bioactive metabolites from Antrodia camphorata ATCC 200183 by asexual reproduction-based repeated batch fermentation. Bioresour. Technol. 2015, 194, 334–343. [Google Scholar] [CrossRef]
- Ren, Y.; Li, H.X.; Zhou, L.; Lu, Z.M.; Shi, J.; Geng, Y.; Xu, Z.H. Protective Effect of Spore Powder of Antrodia camphorata ATCC 200183 on CCl4-Induced Liver Fibrosis in Mice. Nutrients 2020, 12, 2778. [Google Scholar] [CrossRef]
- Zhang, X.L.; Ren, F.; Huang, W.; Ding, R.T.; Zhou, Q.S.; Liu, X.W. Anti-Fatigue Activity of Extracts of Stem Bark from Acanthopanax senticosus. Molecules 2011, 16, 28–37. [Google Scholar] [CrossRef] [Green Version]
- Li-xin, Z.; Xiang-kun, C.; Yu-ping, W.; Di, J.; Peng, D.; Shi-min, T. Study on anti-fatigue function and stability of asparagus extracts in mice. Food Res. Dev. 2021, 42, 37–42. [Google Scholar] [CrossRef]
- Kim, S.; Jo, K.; Byun, B.S.; Han, S.H.; Yu, K.W.; Suh, H.J.; Hong, K.B. Chemical and biological properties of puffed Dendrobium officinale extracts: Evaluation of antioxidant and anti-fatigue activities. J. Funct. Foods 2020, 73, 104144. [Google Scholar] [CrossRef]
- Scheinin, M.; Junnila, J.; Reiner, G.; MacDonald, A.; Muntau, A.C. Nitrogen Balance after the Administration of a Prolonged-Release Protein Substitute for Phenylketonuria as a Single Dose in Healthy Volunteers. Nutrients 2021, 13, 3189. [Google Scholar] [CrossRef]
- Xuan, M.; Hui, C.; Lixing, C.; Shuang, Z.; Chong, Z.; Shutao, Y.; Hongbo, H. Mechanisms of physical fatigue and its applications in nutritional interventions. J. Agric. Food Chem. 2021, 69, 6755–6768. [Google Scholar] [CrossRef]
- Cui, J.; Xia, P.; Zhang, L.; Hu, Y.; Xie, Q.; Xiang, H. A novel fermented soybean, inoculated with selected Bacillus, Lactobacillus and Hansenula strains, showed strong antioxidant and anti-fatigue potential activity. Food Chem. 2020, 333, 127527. [Google Scholar] [CrossRef]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The GenusAlistipes: Gut Bacteria With Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef]
- Fremont, M.; Coomans, D.; Massart, S.; De Meirleir, K. High-throughput 16S rRNA gene sequencing reveals alterations of intestinal microbiota in myalgic encephalomyelitis/chronic fatigue syndrome patients. Anaerobe 2013, 22, 50–56. [Google Scholar] [CrossRef] [Green Version]
- Averina, O.V.; Poluektova, E.U.; Marsova, M.V.; Danilenko, V.N. Biomarkers and Utility of the Antioxidant Potential of Probiotic Lactobacilli and Bifidobacteria as Representatives of the Human Gut Microbiota. Biomedicines 2021, 9, 1340. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Ribbera, A.; Foroni, E.; van Sinderen, D.; Ventura, M. Human gut microbiota and bifidobacteria: From composition to functionality. Antonie Van Leeuwenhoek 2008, 94, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Butterworth, R.F.; Lalonde, R.; Power, C.; Baker, G.B.; Gamrani, H.; Ahboucha, S. Dehydroepiandrosterone sulphate improves cholestasis-associated fatigue in bile duct ligated rats. Neurogastroenterol. Motil. 2009, 21, 1319–1325. [Google Scholar] [CrossRef] [PubMed]
- Fernando, W.; Flint, S.H.; Ranaweera, K.; Bamunuarachchi, A.; Johnson, S.K.; Brennan, C.S. The potential synergistic behaviour of inter- and intra-genus probiotic combinations in the pattern and rate of short chain fatty acids formation during fibre fermentation. Int. J. Food Sci. Nutr. 2018, 69, 144–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuki, T.; Yahagi, K.; Mori, H.; Matsumoto, H.; Hara, T.; Tajima, S.; Ogawa, E.; Kodama, H.; Yamamoto, K.; Yamada, T.; et al. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat. Commun. 2016, 7, 11939. [Google Scholar] [CrossRef]
- Nishiwaki, H.; Hamaguchi, T.; Ito, M.; Ishida, T.; Maeda, T.; Kashihara, K.; Tsuboi, Y.; Ueyama, J.; Shimamura, T.; Mori, H.; et al. Short-Chain Fatty Acid-Producing Gut Microbiota Is Decreased in Parkinson’s Disease but Not in Rapid-Eye-Movement Sleep Behavior Disorder. Msystems 2020, 5, e00797-20. [Google Scholar] [CrossRef]
- Dong, C.R.; Yu, J.Q.; Yang, Y.A.; Zhang, F.; Su, W.Q.; Fan, Q.H.; Wu, C.M.; Wu, S.X. Berberine, a potential prebiotic to indirectly promote Akkermansia growth through stimulating gut mucin secretion. Biomed. Pharmacother. 2021, 139, 111595. [Google Scholar] [CrossRef]
- Woting, A.; Blaut, M. The Intestinal Microbiota in Metabolic Disease. Nutrients 2016, 8, 202. [Google Scholar] [CrossRef]
- Belzer, C.; Chia, L.W.; Aalvink, S.; Chamlagain, B.; Piironen, V.; Knol, J.; Vos, W.M.d.; Dubilier, N. Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and vitamin B12 production by intestinal symbionts. mBio 2017, 8, e00770-17. [Google Scholar] [CrossRef] [Green Version]
- Bu, F.; Zhang, S.H.; Duan, Z.L.; Ding, Y.; Chen, T.; Wang, R.; Feng, Z.Y.; Shi, G.P.; Zhou, J.Y.; Chen, Y.G. A critical review on the relationship of herbal medicine, Akkermansia muciniphila, and human health. Biomed. Pharmacother. 2020, 128, 110352. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, T.; Li, S.; Guan, Q.; Shi, J.-S.; Lu, Z.-M.; Xu, Z.-H.; Geng, Y. Spore Powder of Paecilomyces hepiali Shapes Gut Microbiota to Relieve Exercise-Induced Fatigue in Mice. Nutrients 2022, 14, 2973. https://doi.org/10.3390/nu14142973
Guan T, Li S, Guan Q, Shi J-S, Lu Z-M, Xu Z-H, Geng Y. Spore Powder of Paecilomyces hepiali Shapes Gut Microbiota to Relieve Exercise-Induced Fatigue in Mice. Nutrients. 2022; 14(14):2973. https://doi.org/10.3390/nu14142973
Chicago/Turabian StyleGuan, Tianyue, Shuoshuo Li, Qijie Guan, Jin-Song Shi, Zhen-Ming Lu, Zheng-Hong Xu, and Yan Geng. 2022. "Spore Powder of Paecilomyces hepiali Shapes Gut Microbiota to Relieve Exercise-Induced Fatigue in Mice" Nutrients 14, no. 14: 2973. https://doi.org/10.3390/nu14142973







