Transcriptome Analysis of C. elegans Reveals Novel Targets for DON Cytotoxicity
Abstract
1. Introduction
2. Results
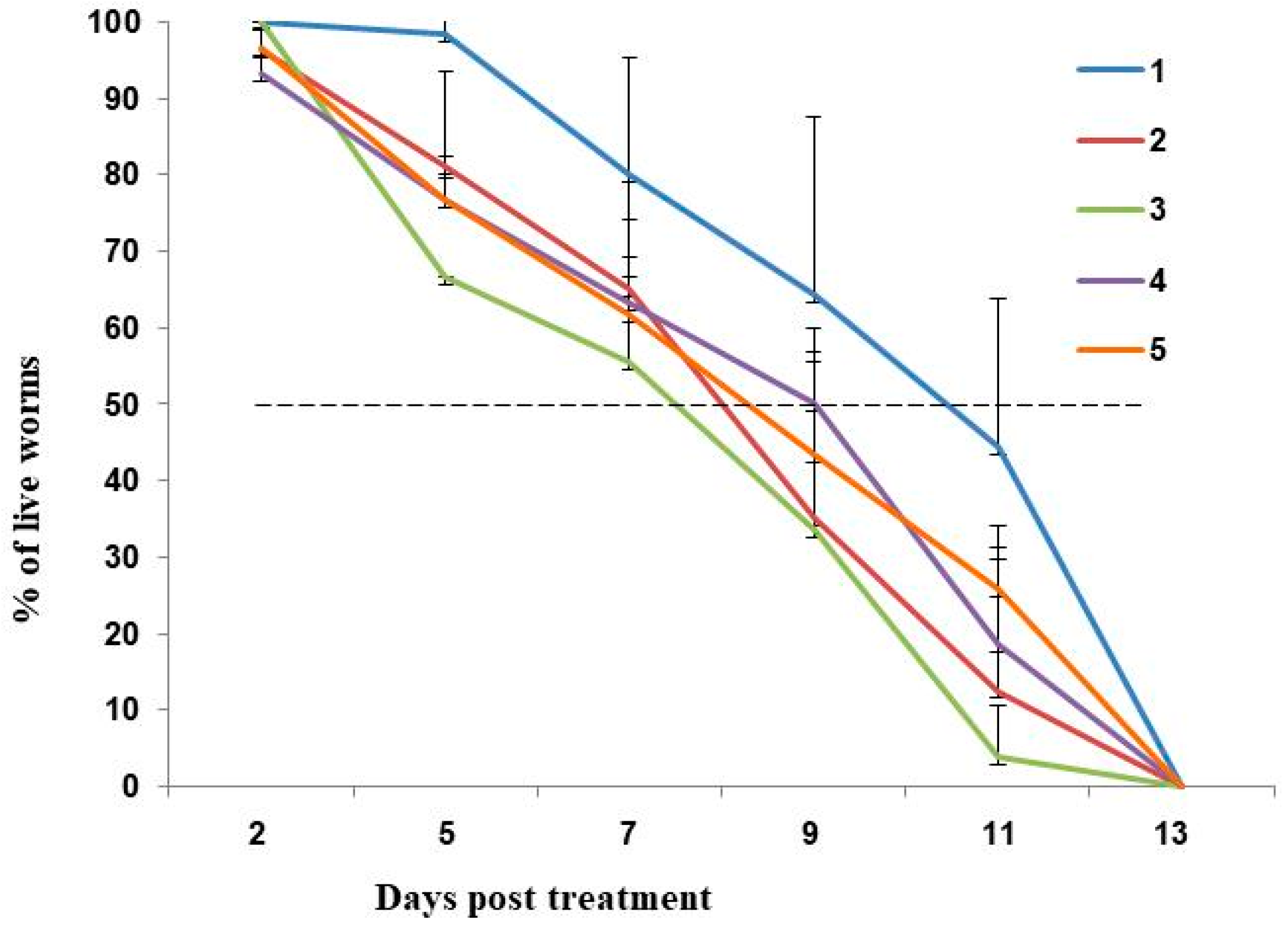
2.1. Deoxynivalenol (DON) Reduces Fecundity and Lifespan of C. elegans
2.2. DON Upregulates Genes Involved in Innate Immunity in C. elegans
2.3. Gene Global Expression is Modulated by DON in RNAseq Analysis
2.4. Functional Analysis of DON-Induced Genes in Conferring DON Tolerance
3. Discussion
4. Materials and Methods
4.1. Materials and Maintenance
4.2. DON Treatment and Lifespan Studies
4.3. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Analysis of Gene Expression
4.4. RNAseq Analysis
4.5. Feeding with RNAi Bacteria
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Di, R.; Blechl, A.; Dill-Macky, R.; Tortora, A.; Tumer, N.E. Expression of a truncated form of yeast ribosomal protein l3 in transgenic wheat improves resistance to fusarium head blight. Plant Sci. 2010, 178, 374–380. [Google Scholar] [CrossRef]
- Wu, F.; Groopman, J.D.; Pestka, J.J. Public health impacts of foodborne mycotoxins. Annu. Rev. Food Sci. Technol. 2014, 5, 351–372. [Google Scholar] [CrossRef] [PubMed]
- Pestka, J.J. Deoxynivalenol: Toxicity, mechanisms and animal health risks. Anim. Feed Sci. Technol. 2007, 137, 283–298. [Google Scholar] [CrossRef]
- Pestka, J.J. Deoxynivalenol: Mechanisms of action, human exposure, and toxicological relevance. Arch. Toxicol. 2010, 84, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Pestka, J.J.; Amuzie, C.J. Tissue distribution and proinflammatory cytokine gene expression following acute oral exposure to deoxynivalenol: Comparison of weanling and adult mice. Food Chem. Toxicol. 2008, 46, 2826–2831. [Google Scholar] [CrossRef] [PubMed]
- Nejdfors, P.; Ekelund, M.; Jeppsson, B.; Westrom, B.R. Mucosal in vitro permeability in the intestinal tract of the pig, the rat, and man: Species- and region-related differences. Scand. J. Gastroenterol. 2000, 35, 501–507. [Google Scholar] [PubMed]
- Szelenyi, I.; Herold, H.; Gothert, M. Emesis induced in domestic pigs: A new experimental tool for detection of antiemetic drugs and for evaluation of emetogenic potential of new anticancer agents. J. Pharmacol. Toxicol. Methods 1994, 32, 109–116. [Google Scholar] [CrossRef]
- Brenner, S. The genetics of dcaenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [PubMed]
- Anderson, G.L.; Boyd, W.A.; Williams, P.L. Assessment of sublethal endpoints for toxicity testing with the nematode caenorhabditis elegans. Environ. Toxicol. Chem. 2001, 20, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Leung, M.C.; Williams, P.L.; Benedetto, A.; Au, C.; Helmcke, K.J.; Aschner, M.; Meyer, J.N. Caenorhabditis elegans: An emerging model in biomedical and environmental toxicology. Toxicol. Sci. 2008, 106, 5–28. [Google Scholar] [CrossRef] [PubMed]
- Boyd, W.A.; McBride, S.J.; Rice, J.R.; Snyder, D.W.; Freedman, J.H. A high-throughput method for assessing chemical toxicity using a caenorhabditis elegans reproduction assay. Toxicol. Appl. Pharmacol. 2010, 245, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Gowrinathan, Y.; Pacan, J.C.; Hawke, A.; Zhou, T.; Sabour, P.M. Toxicity assay for deoxynivalenol using caenorhabditis elegans. Food Addit. Contam. Part A 2011, 28, 1235–1241. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xue, K.S.; Sun, X.; Tang, L.; Wang, J.S. Multi-toxic endpoints of the foodborne mycotoxins in nematode caenorhabditis elegans. Toxins 2015, 7, 5224–5235. [Google Scholar] [CrossRef] [PubMed]
- Parmalee, N.L.; Maqbool, S.B.; Ye, B.; Calder, B.; Bowman, A.B.; Aschner, M. Rnaseq in C. elegans following manganese exposure. Curr. Protoc. Toxicol. 2015, 65. [Google Scholar] [CrossRef]
- Bhaskar, A.S.; Gupta, N.; Rao, P.V. Transcriptomic profile of host response in mouse brain after exposure to plant toxin abrin. Toxicology 2012, 299, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Troemel, E.R.; Chu, S.W.; Reinke, V.; Lee, S.S.; Ausubel, F.M.; Kim, D.H. P38 mapk regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006, 2, e183. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.C.; Chiu, H.C.; Kuo, C.J.; Wu, C.M.; Syu, W.J.; Chiu, W.T.; Chen, C.S. Enterohaemorrhagic escherichia coli o157:H7 shiga-like toxin 1 is required for full pathogenicity and activation of the p38 mitogen-activated protein kinase pathway in caenorhabditis elegans. Cell. Microbiol. 2013, 15, 82–97. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, H.; Bojer, M.S.; Marinus, M.G.; Xu, T.; Struve, C.; Krogfelt, K.A.; Lobner-Olesen, A. The alkaloid compound harmane increases the lifespan of Caenorhabditis elegans during bacterial infection, by modulating the nematode's innate immune response. PLoS ONE 2013, 8, e60519. [Google Scholar] [CrossRef] [PubMed]
- Bock, K.W. The udp-glycosyltransferase (ugt) superfamily expressed in humans, insects and plants: Animal-plant arms-race and co-evolution. Biochem. Pharmacol. 2016, 99, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.; Li, Y.; Lim, E.; Bowles, D.J. Higher plant glycosyltransferases. Genome Biol. 2001, 2, REVIEWS3004. [Google Scholar] [CrossRef] [PubMed]
- Poppenberger, B.; Berthiller, F.; Lucyshyn, D.; Sieberer, T.; Schuhmacher, R.; Krska, R.; Kuchler, K.; Glossl, J.; Luschnig, C.; Adam, G. Detoxification of the fusarium mycotoxin deoxynivalenol by a udp-glucosyltransferase from arabidopsis thaliana. J. Biol. Chem. 2003, 278, 47905–47914. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Torres-Acosta, J.A.; Heinen, S.J.; McCormick, S.; Lemmens, M.; Paris, M.P.; Berthiller, F.; Adam, G.; Muehlbauer, G.J. Transgenic Arabidopsis thaliana expressing a barley udp-glucosyltransferase exhibit resistance to the mycotoxin deoxynivalenol. J. Exp. Bot. 2012, 63, 4731–4740. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shin, S.; Heinen, S.; Dill-Macky, R.; Berthiller, F.; Nersesian, N.; Clemente, T.; McCormick, S.; Muehlbauer, G.J. Transgenic wheat expressing a barley udp-glucosyltransferase detoxifies deoxynivalenol and provides high levels of resistance to fusarium graminearum. Mol. Plant-Microbe Interact. 2015, 28, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- WormBook. Available online: www.wormbook.org (accessed on 1 June 2018).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Nakad, R.; Snoek, L.B.; Yang, W.; Ellendt, S.; Schneider, F.; Mohr, T.G.; Rosingh, L.; Masche, A.C.; Rosenstiel, P.C.; Dierking, K.; et al. Contrasting invertebrate immune defense behaviors caused by a single gene, the Caenorhabditis elegans neuropeptide receptor gene npr-1. BMC Genom. 2016, 17, 280–300. [Google Scholar] [CrossRef] [PubMed]




| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| C17H12.8 | GGCTCCTTGCACTTGTCACA | ATGATTGGCTCAGGGATCTGA |
| K08D8.5 | CCGGGAAGTCGAATGAACAT | GATGCAACACCTGCCAAAGA |
| F35E12.5 | CGCAGTTCAGTTGCCCATT | GGGATTGATCCAGACATTCCA |
| T04C12.6.2 | CTCCACGCGCCGTGTT | CATACCGACCATGACTCCTTGA |
| Gene Symbol | Gene Name by WormBase | Gene Function by WormBase | Fold Change of Gene Expression |
|---|---|---|---|
| F11D11.3 | F11D11.3 gene | unknown | 612.6 + |
| F52F10.4 | oac-32 (o-acyltransferase homolog) | transferase activity, transferring acyl | 513.4 + |
| H23L24.5 | pme-4 [poly(ADP-ribose) metabolism enzyme] | poly(ADP-ribose) glycohydrolase activity | 501.6 + |
| C07G3.2 | irg-1 (infection response gene) | defense response to Gram-negative bacterium, innate immune response | 94.5 + |
| F25A2.1 | F25A2.1 gene | triglyceride lipase activity | 52.0 + |
| Y39G10AR.6 | ugt-31 (UDP-glucuronosyl transferase) | transferase activity, transferring hexosyl groups | 39.5 + |
| T24A6.7 | T24A6.7 gene | unknown | 37.2 + |
| C10H11.6 | ugt-26 (UDP-glucuronosyl transferase) | transferase activity, transferring hexosyl groups | 37.1 + |
| Y49C4A.8 | ugt-29 (UDP-glucuronosyl transferase) | transferase activity, transferring hexosyl groups | 33.1 + |
| C17H12.8 | C17H12.8 gene | PMK-1-regulated gene in innate immunity | 32.4 + |
| T16G1.5 | T16G1.5 gene | transferase activity, transferring phosphorus-containing groups | 28.2 + |
| C10H11.4 | ugt-28 (UDP-glucuronosyl transferase) | transferase activity, transferring hexosyl groups | 27.8 + |
| F08F3.7 | cyp-14A5 (cytochrome P450 family) | heme binding, iron ion binding, oxidoreductase activity | 27.7 + |
| Y58A7A.5 | Y58A7A.5 gene | unknown | 26.7 + |
| Y26D4A.21 | Y26D4A.21 gene | unknown | 25.5 + |
| C49G7.7 | C49G7.7 gene | unknown | 23.7 + |
| Y39A3B.7 | Y39A3B.7 gene | unknown | 23.5 + |
| T05F1.9 | T05F1.9 gene | unknown | 23.3 + |
| F45E4.1 | arf-1.1 (ADP-ribosylation factor related) | GTP binding | 22.4 + |
| F14F9.3 | F14F9.3 gene | zinc ion binding | 21.8 + |
| C06B3.7 | C06B3.7 gene | unknown | 21.5 + |
| F14F9.4 | F14F9.4 gene | zinc ion binding | 20.1 + |
| F22E10.1 | pgp-12 (P-glycoprotein related) | ATP binding, ATPase activity | 18.9 + |
| ZC239.12 | sdz-35 (SKN-1 dependent zygotic transcript) | protein binding | 17.3 + |
| C32H11.1 | C32H11.1 gene (encoding a CUB-like domain containing protine) | innate immune response to several different bacterial pathogens | 17.3 + |
| F14F9.2 | F14F9.2 gene | unknown | 16.7 + |
| Y119D3A.3 | fbxa-35 (F-box A protein) | protein binding | 16.3 + |
| ZK228.4 | ZK228.4 gene | unknown | 15.5 + |
| F56D6.2 | clec-67 (C-type lectin) | carbohydrate binding | 14.9 + |
| F33H12.7 | F33H12.7 gene | unknown | 14.7 + |
| F55G11.5 | dod-22 (nownstream of DAF-16, regulated by DAF-16) | defense response to Gram-negative bacterium, innate immune response | 13.9 + |
| F14H8.6 | cng-1 (cyclic nucleotide gated channel) | ion transmembrane transporter activity | 13.7 + |
| C49G7.5 | irg-2 (infection response gene) | defense response to Gram-negative bacterium, innate immune response | 13.3 + |
| K11D12.5 | swt-7 (SWEET sugar transporter family) | defense response | 13.2 + |
| Y58A7A.3 | Y58A7A.3 gene | zinc ion binding | 12.8 + |
| F43C1.7 | F43C1.7 gene | unknown | 12.7 + |
| C17H12.6 | C17H12.6 gene | unknown | 12.2 + |
| F08E10.7 | scl-24 (SCP-like extracellular protein) | unknown | 12.2 + |
| F36G9.12 | oac-20 (o-acyltransferase homolog) | transferase activity, transferring acyl groups other than amino-acyl groups | 12.2 + |
| Y37H2A.14 | Y37H2A.14 gene | unknown | 12.1 + |
| K08D8.5 | K08D8.5 gene (encoding a CUB-like domain containing protine) | innate immune response to several different bacterial pathogens | 12.0 + |
| F35E12.5 | F35E12.5 gene | unknown | 11.8 + |
| F54D5.4 | F54D5.4 gene | unknown | 10.9 + |
| T05B4.8 | T05B4.8 gene | unknown | 10.2 + |
| W05G11.3 | col-88 (collagen) | structural constituent of cuticle | 415.8 |
| T26H2.5 | sqst-3 (SeQueSTosome related) | zinc ion binding | 250.4 − |
| C42D8.2 | vit-2 (vitellogenin structural genes) (yolk protein genes) | lipid transporter activity | 44.7 − |
| C45G7.3 | ilys-3 (invertebrate lysozyme) | lysozyme activity | 21.3 − |
| F41F3.4 | col-139 (collagen) | structural constituent of cuticle | 17.7 − |
| M18.1 | col-129 (collagen) | structural constituent of cuticle | 16.4 − |
| B0218.8 | clec-52 (C-type lectin) | carbohydrate binding | 15.4 − |
| ZK1193.1 | col-19 (collagen) | structural constituent of cuticle | 12.9 − |
| C36A4.1 | cyp-25A1 (cytochrome P450 family) | heme-binding, iron ion binding, oxidoreductase activity | 12.8 − |
| F26F12.1 | col-140 (collagen) | structural constituent of cuticle | 12.1 − |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di, R.; Zhang, H.; Lawton, M.A. Transcriptome Analysis of C. elegans Reveals Novel Targets for DON Cytotoxicity. Toxins 2018, 10, 262. https://doi.org/10.3390/toxins10070262
Di R, Zhang H, Lawton MA. Transcriptome Analysis of C. elegans Reveals Novel Targets for DON Cytotoxicity. Toxins. 2018; 10(7):262. https://doi.org/10.3390/toxins10070262
Chicago/Turabian StyleDi, Rong, Hanzhong Zhang, and Michael A. Lawton. 2018. "Transcriptome Analysis of C. elegans Reveals Novel Targets for DON Cytotoxicity" Toxins 10, no. 7: 262. https://doi.org/10.3390/toxins10070262
APA StyleDi, R., Zhang, H., & Lawton, M. A. (2018). Transcriptome Analysis of C. elegans Reveals Novel Targets for DON Cytotoxicity. Toxins, 10(7), 262. https://doi.org/10.3390/toxins10070262




