Primary Sequence and 3D Structure Prediction of the Plant Toxin Stenodactylin
Abstract
:1. Introduction
2. Results
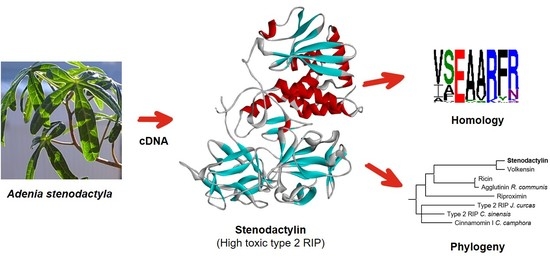
2.1. Stenodactylin Gene Sequence
2.2. Structure of Stenodactylin
2.3. Sequence Comparison between Stenodactylin and Other RIPs
2.4. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Methods
4.2.1. Synthesis of cDNA
4.2.2. cDNA Cloning and Sequence
4.2.3. Sequence Retrieval and Data Treatment
4.2.4. Secondary Structure Prediction
4.2.5. Sequence Alignment
4.2.6. Protein Structure Studies and Graphical Representation
4.2.7. Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bolognesi, A.; Bortolotti, M.; Maiello, S.; Battelli, M.G.; Polito, L. Ribosome-Inactivating Proteins from Plants: A Historical Overview. Molecules 2016, 21, 1627. [Google Scholar] [CrossRef] [PubMed]
- Polito, L.; Bortolotti, M.; Maiello, S.; Battelli, M.G.; Bolognesi, A. Plants Producing Ribosome-Inactivating Proteins in Traditional Medicine. Molecules 2016, 21, 1560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bortolotti, M.; Mercatelli, D.; Polito, L. Momordica charantia, a Nutraceutical Approach for Inflammatory Related Diseases. Front. Pharmacol. 2019, 10, 486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbieri, L.; Valbonesi, P.; Bonora, E.; Gorini, P.; Bolognesi, A.; Stirpe, F. Polynucleotide:adenosine glycosidase activity of ribosome-inactivating proteins: Effect on DNA, RNA and poly(A). Nucleic Acids Res. 1997, 25, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Battelli, M.G.; Barbieri, L.; Bolognesi, A.; Buonamici, L.; Valbonesi, P.; Polito, L.; Van Damme, E.J.M.; Peumans, W.J.; Stirpe, F. Ribosome-inactivating lectins with polynucleotide:adenosine glycosidase activity. FEBS Lett. 1997, 408, 355–359. [Google Scholar] [CrossRef] [Green Version]
- Polito, L.; Bortolotti, M.; Battelli, M.G.; Calafato, G.; Bolognesi, A. Ricin: An Ancient Story for a Timeless Plant Toxin. Toxins 2019, 11, 324. [Google Scholar] [CrossRef] [Green Version]
- Stirpe, F.; Bolognesi, A.; Bortolotti, M.; Farini, V.; Lubelli, C.; Pelosi, E.; Polito, L.; Dozza, B.; Strocchi, P.; Chambery, A.; et al. Characterization of highly toxic type 2 ribosome-inactivating proteins from Adenia lanceolata and Adenia stenodactyla (Passifloraceae). Toxicon 2007, 50, 94–105. [Google Scholar] [CrossRef]
- Wiley, R.G.; Kline, R.H., IV. Neuronal lesioning with axonally transported toxins. J. Neurosci. Methods 2000, 103, 73–82. [Google Scholar] [CrossRef]
- Monti, B.; D’Alessandro, C.; Farini, V.; Bolognesi, A.; Polazzi, E.; Contestabile, A.; Stirpe, F.; Battelli, M.G. In vitro and in vivo toxicity of type 2 ribosome-inactivating proteins lanceolin and stenodactylin on glial and neuronal cells. Neurotoxicology 2007, 28, 637–644. [Google Scholar] [CrossRef]
- Polito, L.; Bortolotti, M.; Pedrazzi, M.; Mercatelli, D.; Battelli, M.G.; Bolognesi, A. Apoptosis and necroptosis induced by stenodactylin in neuroblastoma cells can be completely prevented through caspase inhibition plus catalase or necrostatin-1. Phytomedicine 2016, 23, 32–41. [Google Scholar] [CrossRef] [Green Version]
- Mercatelli, D.; Bortolotti, M.; Andresen, V.; Sulen, A.; Polito, L.; Gjertsen, B.T.; Bolognesi, A. Early response to the plant toxin stenodactylin in acute myeloid leukemia cells involves inflammatory and apoptotic signaling. Front. Pharmacol. 2020, 11, 630. [Google Scholar] [CrossRef] [PubMed]
- Polito, L.; Djemil, A.; Bortolotti, M. Plant Toxin-Based Immunotoxins for Cancer Therapy: A Short Overview. Biomedicines 2016, 4, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puri, M.; Kaur, I.; Perugini, M.A.; Gupta, R.C. Ribosome-inactivating proteins: Current status and biomedical applications. Drug Discov. Today 2012, 17, 774–783. [Google Scholar] [CrossRef] [PubMed]
- De Virgilio, M.; Lombardi, A.; Caliandro, R.; Fabbrini, M.S. Ribosome-inactivating proteins: From plant defense to tumor attack. Toxins 2010, 2, 2699–2737. [Google Scholar] [CrossRef] [Green Version]
- Montfort, W.; Villafranca, J.E.; Monzingo, A.F.; Ernst, S.R.; Katzin, B.; Rutenber, E.; Xuong, N.H.; Hamlin, R.; Robertus, J.D. The three-dimensional structure of ricin at 2.8 A. J. Biol. Chem. 1987, 262, 5398–5403. [Google Scholar]
- Mlsna, D.; Monzingo, A.F.; Katzin, B.J.; Ernst, S.; Robertus, J.D. Structure of recombinant ricin A chain at 2.3 A. Protein Sci. 1993, 2, 429–435. [Google Scholar] [CrossRef] [Green Version]
- Tosi, G.; Fermani, S.; Falini, G.; Polito, L.; Bortolotti, M.; Bolognesi, A. Crystallization and preliminary X-ray diffraction data analysis of stenodactylin, a highly toxic type 2 ribosome-inactivating protein from Adenia stenodactyla. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 51–53. [Google Scholar] [CrossRef] [Green Version]
- ExPASy Bioinformatics Resource Portal. Available online: https://www.expasy.org/ (accessed on 25 June 2020).
- NetNGlyc 1.0 Server. Available online: http://www.cbs.dtu.dk/services/NetNGlyc/ (accessed on 25 June 2020).
- Yan, Q.; Li, X.P.; Tumer, N.E. N-glycosylation does not affect the catalytic activity of ricin A chain but stimulates cytotoxicity by promoting its transport out of the endoplasmic reticulum. Traffic 2012, 13, 1508–1521. [Google Scholar] [CrossRef] [Green Version]
- Sehgal, P.; Kumar, O.; Kameswararao, M.; Ravindran, J.; Khan, M.; Sharma, S.; Vijayaraghavan, R.; Prasad, G.B.K.S. Differential toxicity profile of ricin isoforms correlates with their glycosylation levels. Toxicology 2011, 282, 56–67. [Google Scholar] [CrossRef]
- Buchan, D.W.A.; Jones, D.T. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 2019, 47, W402–W407. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Rutenber, E.; Katzin, B.J.; Ernst, S.; Collins, E.J.; Mlsna, D.; Ready, M.P.; Robertus, J.D. Crystallographic refinement of ricin to 2.5 A. Proteins 1991, 10, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Tahirov, T.H.; Lu, T.H.; Liaw, Y.C.; Chen, Y.L.; Lin, J.Y. Crystal structure of abrin-a at 2.14 A. J. Mol. Biol. 1995, 250, 354–367. [Google Scholar] [CrossRef] [PubMed]
- Di Maro, A.; Citores, L.; Russo, R.; Iglesias, R.; Ferreras, J.M. Sequence comparison and phylogenetic analysis by the Maximum Likelihood method of ribosome-inactivating proteins from angiosperms. Plant Mol. Biol. 2014, 85, 575–588. [Google Scholar] [CrossRef]
- Deeks, E.D.; Cook, J.P.; Day, P.J.; Smith, D.C.; Roberts, L.M.; Lord, J.M. The low lysine content of ricin a chain reduces the risk of proteolytic degradation after translocation from the endoplasmic reticulum to the cytosol. Biochemistry 2002, 41, 3405–3413. [Google Scholar] [CrossRef]
- Katzin, B.J.; Collins, E.J.; Robertus, J.D. Structure of ricin A-chain at 2.5 A. Proteins 1991, 10, 251–259. [Google Scholar] [CrossRef]
- Monzingo, A.F.; Robertus, J.D. X-ray analysis of substrate analogs in the ricin A-chain active site. J. Mol. Biol. 1992, 227, 1136–1145. [Google Scholar] [CrossRef]
- Hao, Q.; Van Damme, E.J.; Hause, B.; Barre, A.; Chen, Y.; Rougé, P.; Peumans, W.J. Iris bulbs express type 1 and type 2 ribosome-inactivating proteins with unusual properties. Plant Physiol. 2001, 125, 866–876. [Google Scholar] [CrossRef] [Green Version]
- Rutenber, E.; Robertus, J.D. Structure of ricin B-chain at 2.5 A resolution. Proteins 1991, 10, 260–269. [Google Scholar] [CrossRef]
- Chambery, A.; Di Maro, A.; Monti, M.M.; Stirpe, F.; Parente, A. Volkensin from Adenia volkensii Harms (kilyambiti plant), a type 2 ribosome-inactivating protein. Eur. J. Biochem. 2004, 271, 108–117. [Google Scholar] [CrossRef]
- Roberts, L.M.; Lamb, F.I.; Pappin, D.J.; Lord, J.M. The primary sequence of Ricinus communis agglutinin. Comparison with ricin. J. Biol. Chem. 1985, 260, 15682–15686. [Google Scholar] [PubMed]
- Van Damme, E.J.; Hao, Q.; Charels, D.; Barre, A.; Rougé, P.; Van Leuven, F.; Peumans, W.J. Characterization and molecular cloning of two different type 2 ribosome-inactivating proteins from the monocotyledonous plant Polygonatum multiflorum. Eur. J. Biochem. 2000, 267, 2746–2759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehar, S.M.; Pedersen, J.T.; Kamath, R.S.; Swimmer, C.; Goldmacher, V.S.; Lambert, J.M.; Blättler, W.A.; Guild, B.C. Mutational and structural analysis of the lectin activity in binding domain 2 of ricin B chain. Protein Eng. 1994, 7, 1261–1266. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.Z.; Zou, W.G.; Liu, W.Y.; Liu, X.Y. The lower cytotoxicity of cinnamomin (a type II RIP) is due to its B-chain. Arch. Biochem. Biophys. 2006, 451, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Pascal, J.M.; Day, P.J.; Monzingo, A.F.; Ernst, S.R.; Robertus, J.D.; Iglesias, R.; Pérez, Y.; Ferreras, J.M.; Citores, L.; Girbés, T. 2.8-A crystal structure of a nontoxic type-II ribosome-inactivating protein, ebulin l. Proteins 2001, 43, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Ferreras, J.M.; Citores, L.; Iglesias, R.; Jiménez, P.; Girbés, T. Sambucus Ribosome-Inactivating Proteins and Lectins. Toxic Plant Proteins. In Toxic Plant Proteins–Plant Cell Monographs, 1st ed.; Lord, J.M., Hartley, M.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 18, pp. 107–131. [Google Scholar] [CrossRef]
- Iglesias, R.; Ferreras, J.M.; Di Maro, A.; Citores, L. Ebulin-RP, a novel member of the Ebulin gene family with low cytotoxicity as a result of deficient sugar binding domains. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 460–473. [Google Scholar] [CrossRef]
- Barbieri, L.; Battelli, M.G.; Stirpe, F. Ribosome-inactivating proteins from plants. Biochim. Biophys. Acta. 1993, 1154, 237–282. [Google Scholar] [CrossRef]
- Villafranca, J.E.; Robertus, J.D. Ricin B chain is a product of gene duplication. J. Biol. Chem. 1981, 256, 554–556. [Google Scholar]
- Blast-Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 25 June 2020).
- Rojo, M.A.; Yato, M.; Ishii-Minami, N.; Minami, E.; Kaku, H.; Citores, L.; Girbés, T.; Shibuya, N. Isolation, cDNA cloning, biological properties, and carbohydrate binding specificity of sieboldin-b, a type II ribosome-inactivating protein from the bark of japanese elderberry (Sambucus sieboldiana). Arch. Biochem. Biophys. 1997, 340, 185–194. [Google Scholar] [CrossRef]
- Kaku, H.; Tanaka, Y.; Tazaki, K.; Minami, E.; Mizuno, H.; Shibuya, N. Sialylated oligosaccharide-specific plant lectin from japanese elderberry (Sambucus sieboldiana) bark tissue has a homologous structure to type II ribosome-inactivating proteins, ricin and abrin: CDNA cloning and molecular modeling study. J. Biol. Chem. 1996, 271, 1480–1485. [Google Scholar] [CrossRef] [Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Nordahl Petersen, T.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.D.; Stephens, R.M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [Green Version]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]








| RIP Name | Identity (%) Stenodactylin A Chain | Identity (%) Stenodactylin B Chain | Identity (%) Stenodactylin Whole Molecule | |
|---|---|---|---|---|
| Type 2 | Volkensin | 81.7 | 90.3 | 86.1 |
| Ricin | 31.4 | 47.7 | 40.3 | |
| Viscumin | 31.0 | 46.0 | 38.1 | |
| Abrin a | 31.2 | 44.6 | 37.9 | |
| Riproximin | 27.7 | 43.3 | 35.8 | |
| Cinnamomin | 33.0 | 42.1 | 37.3 | |
| Ebulin l | 28.7 | 44.2 | 36.5 | |
| Nigrin b | 29.0 | 43.9 | 36.5 | |
| Type 1 | Saporin | 18.9 | ||
| Dianthin | 18.1 | |||
| Momordin | 24.0 |
| Primer | Sequence |
|---|---|
| STA2 | 5′ GCCACGGTAGAGAGRTACACT 3′ |
| STB1R | 5′ AAGTCGTCTCCCCGGAAGGGC 3′ |
| STB3R | 5′ GGCGGGGTTGATGGTTCC 3′ |
| STB1 | 5′ TGCCCTTCCGGGGAGACGACT 3′ |
| STB5R | 5′ TAGGAACCATTGCTGGTTGGA 3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iglesias, R.; Polito, L.; Bortolotti, M.; Pedrazzi, M.; Citores, L.; Ferreras, J.M.; Bolognesi, A. Primary Sequence and 3D Structure Prediction of the Plant Toxin Stenodactylin. Toxins 2020, 12, 538. https://doi.org/10.3390/toxins12090538
Iglesias R, Polito L, Bortolotti M, Pedrazzi M, Citores L, Ferreras JM, Bolognesi A. Primary Sequence and 3D Structure Prediction of the Plant Toxin Stenodactylin. Toxins. 2020; 12(9):538. https://doi.org/10.3390/toxins12090538
Chicago/Turabian StyleIglesias, Rosario, Letizia Polito, Massimo Bortolotti, Manuela Pedrazzi, Lucía Citores, José M. Ferreras, and Andrea Bolognesi. 2020. "Primary Sequence and 3D Structure Prediction of the Plant Toxin Stenodactylin" Toxins 12, no. 9: 538. https://doi.org/10.3390/toxins12090538