Dramatically Enhancing the Sensitivity of Immunoassay for Ochratoxin A Detection by Cascade-Amplifying Enzyme Loading
Abstract
:1. Introduction
2. Results and Discussion
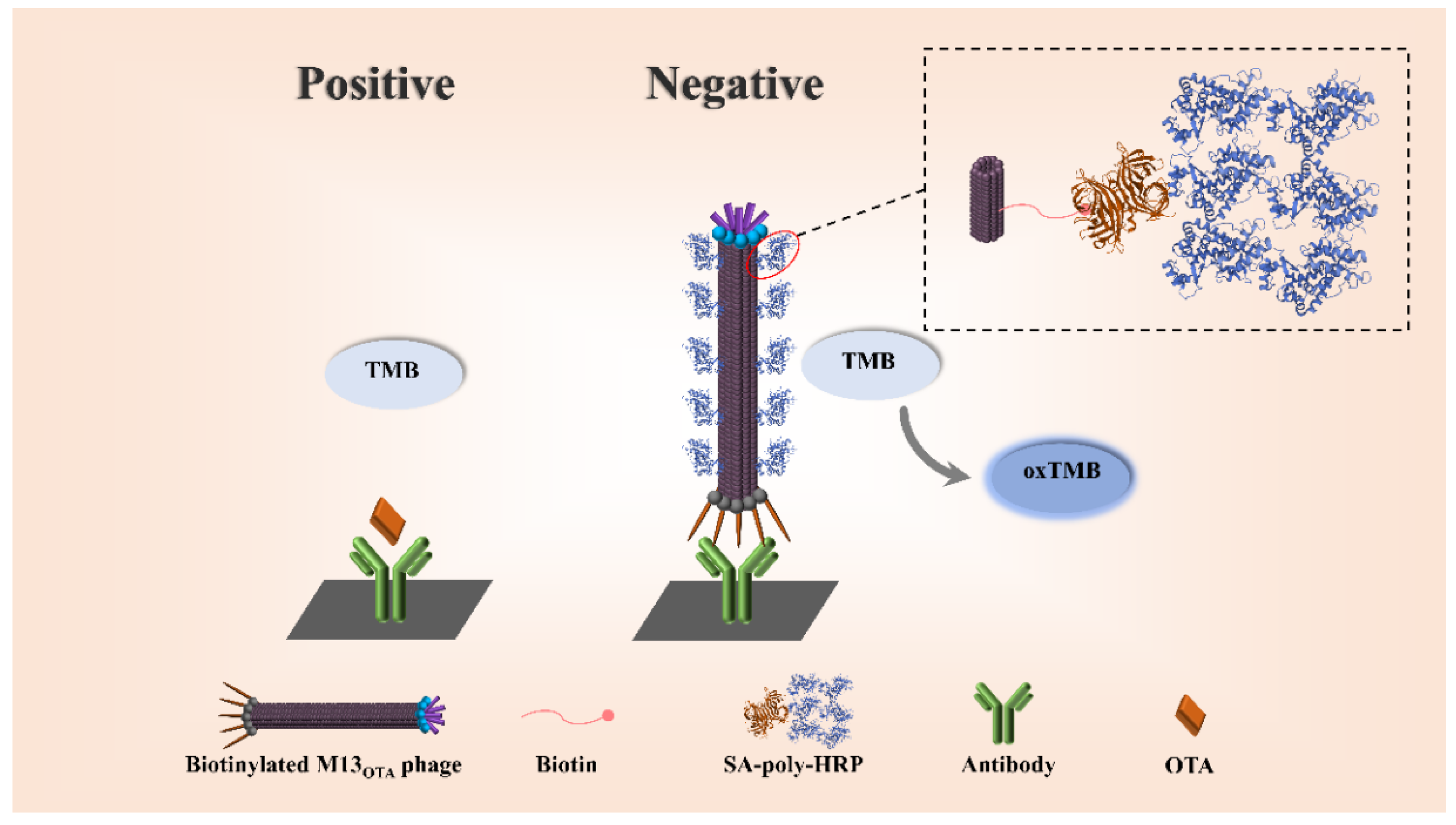
2.1. Principle of the Proposed Bio-M13OTA-ELISA Method
2.2. Preparation of Bio-M13OTA Phage
2.3. Development of Bio-M13OTA-ELISA
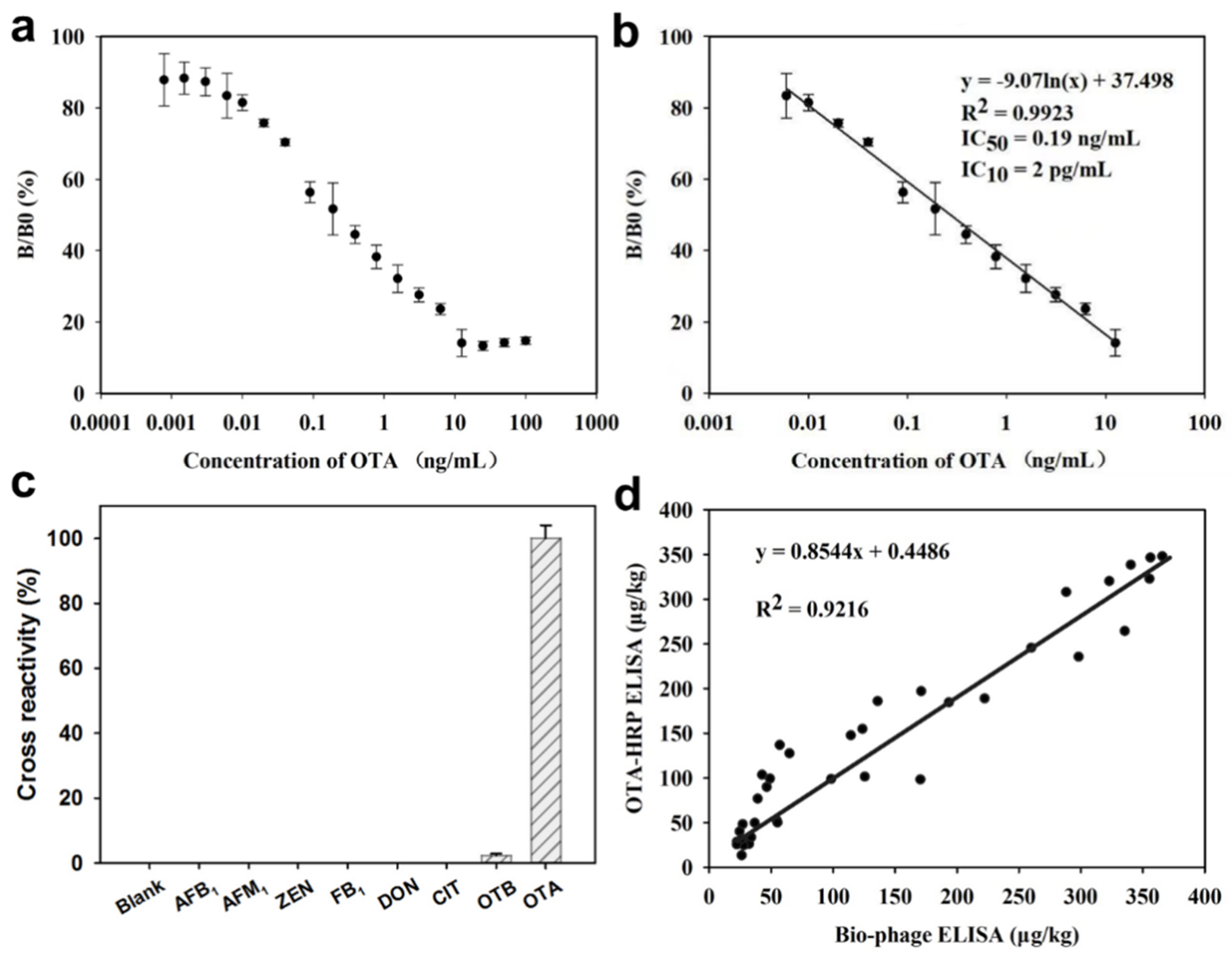
2.4. Validation of Analytical Performance
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Propagation of M13OTA Bacteriophage
4.3. Preparation of Biotinylated M13OTA (Bio-M13OTA) Bacteriophage
4.4. Assay Procedure of Bio-M13OTA-ELISA for OTA Detection
4.5. Detection Performance of the Proposed Bio-M13OTA-ELISA for OTA Detection
4.6. Assay Procedure of Conventional HRP Based-ELISA for OTA Detection
4.7. Corn Sample Pretreatment
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van Der Merwe, K.J.; Steyn, P.S.; Fourie, L.; Scott, D.B.; Theron, J.J. Ochratoxin A, a Toxic Metabolite produced by Aspergillus ochraceus Wilh. Nature 1965, 205, 1112–1113. [Google Scholar] [CrossRef] [PubMed]
- Fink-Gremmels, J.; Jahn, A.; Blom, M.J. Toxicity and metabolism of ochratoxin A. Nat. Toxins 1995, 3, 214–220. [Google Scholar] [CrossRef]
- Reddy, K.R.N.; Salleh, B.; Saad, B.; Abbas, H.K.; Abel, C.A.; Shier, W.T. An overview of mycotoxin contamination in foods and its implications for human health. Toxin Rev. 2010, 29, 3–26. [Google Scholar] [CrossRef]
- Duarte, S.C.; Lino, C.M.; Pena, A. Mycotoxin food and feed regulation and the specific case of ochratoxin A: A review of the worldwide status. Food Addit. Contam. Part A 2010, 27, 1440–1450. [Google Scholar] [CrossRef]
- Yang, L.; Wang, L.; Pan, J.; Xiang, L.; Yang, M.; Logrieco, A.F. Determination of ochratoxin A in traditional Chinese medicinal plants by HPLC–FLD. Food Addit. Contam. Part A 2010, 27, 989–997. [Google Scholar] [CrossRef]
- Al-Taher, F.; Banaszewski, K.; Jackson, L.; Zweigenbaum, J.; Ryu, D.; Cappozzo, J. Rapid Method for the Determination of Multiple Mycotoxins in Wines and Beers by LC-MS/MS Using a Stable Isotope Dilution Assay. J. Agric. Food Chem. 2013, 61, 2378–2384. [Google Scholar] [CrossRef]
- Zhang, X.; Li, M.; Cheng, Z.; Ma, L.; Zhao, L.; Li, J. A comparison of electronic nose and gas chromatography–mass spectrometry on discrimination and prediction of ochratoxin A content in Aspergillus carbonarius cultured grape-based medium. Food Chem. 2019, 297, 124850. [Google Scholar] [CrossRef]
- Duan, H.; Huang, X.; Shao, Y.; Zheng, L.; Guo, L.; Xiong, Y. Size-Dependent Immunochromatographic Assay with Quantum Dot Nanobeads for Sensitive and Quantitative Detection of Ochratoxin A in Corn. Anal. Chem. 2017, 89, 7062–7068. [Google Scholar] [CrossRef]
- Flajs, D.; Domijan, A.M.; Ivić, D.; Cvjetković, B.; Peraica, M. ELISA and HPLC analysis of ochratoxin A in red wines of Croatia. Food Control 2009, 20, 590–592. [Google Scholar] [CrossRef]
- Xiong, Y.; Leng, Y.; Li, X.; Huang, X.; Xiong, Y. Emerging strategies to enhance the sensitivity of competitive ELISA for detection of chemical contaminants in food samples. TrAC Trends Anal. Chem. 2020, 126, 115861. [Google Scholar] [CrossRef]
- Pei, K.; Xiong, Y.; Xu, B.; Wu, K.; Li, X.; Jiang, H.; Xiong, Y. Colorimetric ELISA for ochratoxin A detection based on the urease-induced metallization of gold nanoflowers. Sens. Actuators B Chem. 2018, 262, 102–109. [Google Scholar] [CrossRef]
- Wang, Y.; Li, P.; Majkova, Z.; Bever, C.R.S.; Kim, H.J.; Zhang, Q.; Dechant, J.E.; Gee, S.J.; Hammock, B.D. Isolation of Alpaca Anti-Idiotypic Heavy-Chain Single-Domain Antibody for the Aflatoxin Immunoassay. Anal. Chem. 2013, 85, 8298–8303. [Google Scholar] [CrossRef] [Green Version]
- Guan, D.; Li, P.; Cui, Y.; Zhang, Q.; Zhang, W. A competitive immunoassay with a surrogate calibrator curve for aflatoxin M1 in milk. Anal. Chim. Acta 2011, 703, 64–69. [Google Scholar] [CrossRef]
- Shu, M.; Xu, Y.; Wang, D.; Liu, X.; Li, Y.; He, Q.; Tu, Z.; Qiu, Y.; Ji, Y.; Wang, X. Anti-idiotypic nanobody: A strategy for development of sensitive and green immunoassay for Fumonisin B1. Talanta 2015, 143, 388–393. [Google Scholar] [CrossRef]
- Hou, S.-L.; Ma, Z.-E.; Meng, H.; Xu, Y.; He, Q.-H. Ultrasensitive and green electrochemical immunosensor for mycotoxin ochratoxin A based on phage displayed mimotope peptide. Talanta 2019, 194, 919–924. [Google Scholar] [CrossRef]
- Yuan, Q.; Pestka, J.J.; Hespenheide, B.M.; Kuhn, L.A.; Linz, J.E.; Hart, L.P. Identification of Mimotope Peptides Which Bind to the Mycotoxin Deoxynivalenol-Specific Monoclonal Antibody. Appl. Environ. Microbiol. 1999, 65, 3279–3286. [Google Scholar] [CrossRef] [Green Version]
- Zou, X.; Chen, C.; Huang, X.; Chen, X.; Wang, L.; Xiong, Y. Phage-free peptide ELISA for ochratoxin A detection based on biotinylated mimotope as a competing antigen. Talanta 2016, 146, 394–400. [Google Scholar] [CrossRef]
- He, Q.-H.; Xu, Y.; Huang, Y.-H.; Liu, R.-R.; Huang, Z.-B.; Li, Y.-P. Phage-displayed peptides that mimic zearalenone and its application in immunoassay. Food Chem. 2011, 126, 1312–1315. [Google Scholar] [CrossRef]
- Liu, X.; Xu, Y.; He, Q.-H.; He, Z.-Y.; Xiong, Z.-P. Application of Mimotope Peptides of Fumonisin B1 in Peptide ELISA. J. Agric. Food Chem. 2013, 61, 4765–4770. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Y.; Gao, X.; Duan, Z.; Wang, S. Determination of Chloramphenicol Residues in Milk by Enzyme-Linked Immunosorbent Assay: Improvement by Biotin−Streptavidin-Amplified System. J. Agric. Food Chem. 2010, 58, 3265–3270. [Google Scholar] [CrossRef]
- Molek, P.; Bratkovič, T. Bacteriophages as Scaffolds for Bipartite Display: Designing Swiss Army Knives on a Nanoscale. Bioconjugate Chem. 2015, 26, 367–378. [Google Scholar] [CrossRef]
- Wang, X.S.; Chen, P.-H.C.; Hampton, J.T.; Tharp, J.M.; Reed, C.A.; Das, S.K.; Wang, D.-S.; Hayatshahi, H.S.; Shen, Y.; Liu, J.; et al. A Genetically Encoded, Phage-Displayed Cyclic-Peptide Library. Angew. Chem. Int. Ed. 2019, 58, 15904–15909. [Google Scholar] [CrossRef]
- Hua, X.; Zhou, L.; Feng, L.; Ding, Y.; Shi, H.; Wang, L.; Gee, S.J.; Hammock, B.D.; Wang, M. Competitive and noncompetitive phage immunoassays for the determination of benzothiostrobin. Anal. Chim. Acta 2015, 890, 150–156. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Jiang, D.-J.; Yan, J.-X.; Ma, Z.-E.; Luo, X.-E.; Wei, T.-L.; Xu, Y.; He, Q.-H. An ultrasensitive electrochemical immunosensor for Cry1Ab based on phage displayed peptides. Talanta 2018, 179, 646–651. [Google Scholar] [CrossRef]
- Lentini, G.; Fazio, E.; Calabrese, F.; De Plano, L.M.; Puliafico, M.; Franco, D.; Nicolò, M.S.; Carnazza, S.; Trusso, S.; Allegra, A.; et al. Phage–AgNPs complex as SERS probe for U937 cell identification. Biosens. Bioelectron. 2015, 74, 398–405. [Google Scholar] [CrossRef]
- Liu, P.; Han, L.; Wang, F.; Petrenko, V.A.; Liu, A. Gold nanoprobe functionalized with specific fusion protein selection from phage display and its application in rapid, selective and sensitive colorimetric biosensing of Staphylococcus aureus. Biosens. Bioelectron. 2016, 82, 195–203. [Google Scholar] [CrossRef]
- Blaik, R.A.; Lan, E.; Huang, Y.; Dunn, B. Gold-Coated M13 Bacteriophage as a Template for Glucose Oxidase Biofuel Cells with Direct Electron Transfer. ACS Nano 2016, 10, 324–332. [Google Scholar] [CrossRef]
- Fang, H.; Zhan, S.; Feng, L.; Chen, X.; Guo, Q.; Guo, Y.; He, Q.; Xiong, Y. Chemical modification of M13 bacteriophage as nanozyme container for dramatically enhanced sensitivity of colorimetric immunosensor. Sens. Actuators B Chem. 2021, 346, 130368. [Google Scholar] [CrossRef]
- Wang, L.; Xiong, S.; Zou, X.; Chen, C.; Shao, H.; Chen, X. Mimic Epitope of Ochratoxin A and Its Application in Phage Enzyme-Linked Immunosorbent Assay. Chin. J. Anal. Chem. 2015, 43, 856–861. [Google Scholar]
- Li, D.; Ying, Y.; Wu, J.; Niessner, R.; Knopp, D. Comparison of monomeric and polymeric horseradish peroxidase as labels in competitive ELISA for small molecule detection. Microchim. Acta 2013, 180, 711–717. [Google Scholar] [CrossRef]
- Hu, J.; Zhou, S.; Zeng, L.; Chen, Q.; Duan, H.; Chen, X.; Li, X.; Xiong, Y. Hydrazide mediated oriented coupling of antibodies on quantum dot beads for enhancing detection performance of immunochromatographic assay. Talanta 2021, 223, 121723. [Google Scholar] [CrossRef] [PubMed]


| OTA Added (μg/kg) | Intra-Assay Precision a | Inter-Assay Precision b | ||||
|---|---|---|---|---|---|---|
| Mean (µg/kg) | Recovery (%) | RSD (%) | Mean (µg/kg) | Recovery (%) | RSD (%) | |
| 50 | 50.83 ± 0.04 | 101.66 | 5.85 | 50.73 ± 0.03 | 101.46 | 5.40 |
| 25 | 24.81 ± 0.03 | 99.27 | 4.10 | 24.45 ± 0.10 | 97.83 | 13.18 |
| 20 | 21.84 ± 0.07 | 109.23 | 9.86 | 20.15 ± 0.05 | 100.76 | 6.47 |
| 10 | 10.36 ± 0.05 | 103.60 | 5.67 | 10.14 ± 0.04 | 101.46 | 5.09 |
| 5 | 5.08 ± 0.09 | 101.67 | 9.91 | 5.04 ± 0.06 | 100.81 | 6.87 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Z.; Feng, L.; Leng, Y.; Huang, M.; Fang, H.; Tong, W.; Chen, X.; Xiong, Y. Dramatically Enhancing the Sensitivity of Immunoassay for Ochratoxin A Detection by Cascade-Amplifying Enzyme Loading. Toxins 2021, 13, 781. https://doi.org/10.3390/toxins13110781
Song Z, Feng L, Leng Y, Huang M, Fang H, Tong W, Chen X, Xiong Y. Dramatically Enhancing the Sensitivity of Immunoassay for Ochratoxin A Detection by Cascade-Amplifying Enzyme Loading. Toxins. 2021; 13(11):781. https://doi.org/10.3390/toxins13110781
Chicago/Turabian StyleSong, Zhuolin, Lin Feng, Yuankui Leng, Mingzhu Huang, Hao Fang, Weipeng Tong, Xuelan Chen, and Yonghua Xiong. 2021. "Dramatically Enhancing the Sensitivity of Immunoassay for Ochratoxin A Detection by Cascade-Amplifying Enzyme Loading" Toxins 13, no. 11: 781. https://doi.org/10.3390/toxins13110781





