Cellular Activity of Salmonella Typhimurium ArtAB Toxin and Its Receptor-Binding Subunit
Abstract
:1. Introduction
2. Results
2.1. Phylogenetic Analysis of AB5 Toxin Subunits

2.2. Purification and Binding of ArtABHIS
2.3. Cloning and Glycan Affinity Purification of ArtAB and ArtB
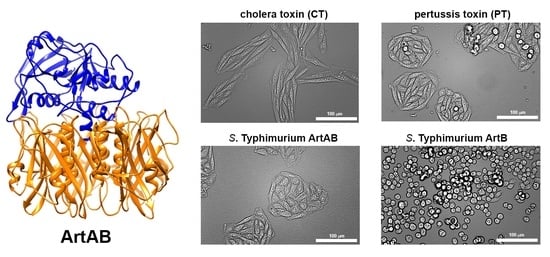
2.4. Cellular Activity of ArtAB
2.5. Cellular Activity of ArtB
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Strains and Growth Conditions
5.2. Phylogenetic Analysis and Predicted Tertiary Structure of AB5 Toxin Subunits
5.3. Construction of ArtABHIS, ArtAB, and ArtB Expression Plasmids
5.4. Expression and Purification of ArtABHIS, ArtAB, and ArtB from E. coli
5.5. ELISA Binding Assays
5.6. Cell Culture
5.7. Fluorescence Microscopy
5.8. Cellular Activity and Morphology Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tack, D.M.; Ray, L.; Griffin, P.M.; Cieslak, P.R.; Dunn, J.; Rissman, T.; Jervis, R.; Lathrop, S.; Muse, A.; Duwell, M.; et al. Preliminary Incidence and Trends of Infections with Pathogens Transmitted Commonly Through Food—Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2016–2019. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 509–514. [Google Scholar] [CrossRef]
- World Health Organization; Food and Agriculture Organization of the United Nations. Interventions for the Control of Non-Typhoidal Salmonella spp. in Beef and Pork: Meeting Report and Systematic Review; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- CDC. Antibiotic Resistance Threats in the United States; U.S. Department of Health and Human Services: Washington, DC, USA, 2019. [Google Scholar]
- Knodler, L.A.; Elfenbein, J.R. Salmonella enterica. Trends Microbiol. 2019, 27, 964–965. [Google Scholar] [CrossRef]
- Morningstar-Shaw, B.R.; Mackie, B.S.A.; Barker, D.K.; Palmer, E.A. Salmonella Serotypes Isolated from Animals and Related Sources; National Center for Emerging and Zoonotic Infectious Diseases, Division of Foodborne, Waterborne, and Environmental Disease, CDC: Atlanta, GA, USA, 2016. [Google Scholar]
- Cheng, R.A.; Eade, C.R.; Wiedmann, M. Embracing Diversity: Differences in Virulence Mechanisms, Disease Severity, and Host Adaptations Contribute to the Success of Nontyphoidal. Front Microbiol. 2019, 10, 1368. [Google Scholar] [CrossRef] [Green Version]
- Leekitcharoenphon, P.; Hendriksen, R.S.; Le Hello, S.; Weill, F.X.; Baggesen, D.L.; Jun, S.R.; Ussery, D.W.; Lund, O.; Crook, D.W.; Wilson, D.J.; et al. Global Genomic Epidemiology of Salmonella enterica Serovar Typhimurium DT104. Appl. Environ. Microbiol. 2016, 82, 2516–2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, C.A.; Fedorka-Cray, P.J.; Vazquez-Torres, A.; Suyemoto, M.; Altier, C.; Ryder, L.R.; Fang, F.C.; Libby, S.J. In vitro and in vivo assessment of Salmonella enterica serovar Typhimurium DT104 virulence. Infect. Immun. 2001, 69, 4673–4677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poppe, C.; Smart, N.; Khakhria, R.; Johnson, W.; Spika, J.; Prescott, J. Salmonella Typhimurium DT104: A virulent and drug-resistant pathogen. Can. Vet. J. 1998, 39, 559–565. [Google Scholar] [PubMed]
- Saitoh, M.; Tanaka, K.; Nishimori, K.; Makino, S.; Kanno, T.; Ishihara, R.; Hatama, S.; Kitano, R.; Kishima, M.; Sameshima, T.; et al. The artAB genes encode a putative ADP-ribosyltransferase toxin homologue associated with Salmonella enterica serovar Typhimurium DT104. Microbiology 2005, 151, 3089–3096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guichard, A.; Moreno, B.C.; Aguilar, B.; van Sorge, N.M.; Kuang, J.; Kurkciyan, A.A.; Wang, Z.; Hang, S.; Pineton de Chambrun, G.P.; McCole, D.F.; et al. Cholera toxin disrupts barrier function by inhibiting exocyst-mediated trafficking of host proteins to intestinal cell junctions. Cell Host. Microbe. 2013, 14, 294–305. [Google Scholar] [CrossRef] [Green Version]
- Kügler, S.; Böcker, K.; Heusipp, G.; Greune, L.; Kim, K.S.; Schmidt, M.A. Pertussis toxin transiently affects barrier integrity, organelle organization and transmigration of monocytes in a human brain microvascular endothelial cell barrier model. Cell Microbiol. 2007, 9, 619–632. [Google Scholar] [CrossRef]
- Cheng, R.A.; Wiedmann, M. The ADP-Ribosylating Toxins of Salmonella. Toxins 2019, 11, 416. [Google Scholar] [CrossRef] [Green Version]
- Tamamura, Y.; Tanaka, K.; Uchida, I. Characterization of pertussis-like toxin from Salmonella spp. that catalyzes ADP-ribosylation of G proteins. Sci. Rep. 2017, 7, 2653. [Google Scholar] [CrossRef] [Green Version]
- Uchida, I.; Ishihara, R.; Tanaka, K.; Hata, E.; Makino, S.; Kanno, T.; Hatama, S.; Kishima, M.; Akiba, M.; Watanabe, A.; et al. Salmonella enterica serotype Typhimurium DT104 ArtA-dependent modification of pertussis toxin-sensitive G proteins in the presence of [32P]NAD. Microbiology 2009, 155, 3710–3718. [Google Scholar] [CrossRef] [Green Version]
- Lencer, W.I.; Constable, C.; Moe, S.; Rufo, P.A.; Wolf, A.; Jobling, M.G.; Ruston, S.P.; Madara, J.L.; Holmes, R.K.; Hirst, T.R. Proteolytic activation of cholera toxin and Escherichia coli labile toxin by entry into host epithelial cells. Signal transduction by a protease-resistant toxin variant. J. Biol. Chem. 1997, 272, 15562–15568. [Google Scholar] [CrossRef] [Green Version]
- Millen, S.H.; Lewallen, D.M.; Herr, A.B.; Iyer, S.S.; Weiss, A.A. Identification and characterization of the carbohydrate ligands recognized by pertussis toxin via a glycan microarray and surface plasmon resonance. Biochemistry 2010, 49, 5954–5967. [Google Scholar] [CrossRef] [Green Version]
- Cabrera, A.; Lepage, J.E.; Sullivan, K.M.; Seed, S.M. Vaxchora: A Single-Dose Oral Cholera Vaccine. Ann. Pharmacother. 2017, 51, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Gregg, K.A.; Merkel, T.J. Pertussis Toxin: A Key Component in Pertussis Vaccines? Toxins 2019, 11, 557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czerkinsky, C.; Sun, J.B.; Lebens, M.; Li, B.L.; Rask, C.; Lindblad, M.; Holmgren, J. Cholera toxin B subunit as transmucosal carrier-delivery and immunomodulating system for induction of antiinfectious and antipathological immunity. Ann. N. Y. Acad. Sci. 1996, 778, 185–193. [Google Scholar] [CrossRef]
- Adkins, I.; Holubova, J.; Kosova, M.; Sadilkova, L. Bacteria and their toxins tamed for immunotherapy. Curr. Pharm. Biotechnol. 2012, 13, 1446–1473. [Google Scholar] [CrossRef]
- Goletz, T.J.; Klimpel, K.R.; Leppla, S.H.; Keith, J.M.; Berzofsky, J.A. Delivery of antigens to the MHC class I pathway using bacterial toxins. Hum. Immunol. 1997, 54, 129–136. [Google Scholar] [CrossRef]
- Morón, G.; Dadaglio, G.; Leclerc, C. New tools for antigen delivery to the MHC class I pathway. Trends Immunol. 2004, 25, 92–97. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Littler, D.R.; Ang, S.Y.; Moriel, D.G.; Kocan, M.; Kleifeld, O.; Johnson, M.D.; Tran, M.T.; Paton, A.W.; Paton, J.C.; Summers, R.J.; et al. Structure-function analyses of a pertussis-like toxin from pathogenic E. coli reveal a distinct mechanism of inhibition of timeric G-proteins. J. Biol. Chem. 2017, 292, 15143–15158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souvorov, A.; Agarwala, R.; Lipman, D.J. SKESA: Strategic k-mer extension for scrupulous assemblies. Genome Biol. 2018, 19, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Gao, X.; Galán, J.E. Structure and function of the Salmonella Typhi chimaeric A2B5 typhoid toxin. Nature 2013, 499, 350–354. [Google Scholar] [CrossRef]
- Gaballa, A.; Cheng, R.A.; Harrand, A.S.; Cohn, A.R.; Wiedmann, M. The Majority of Typhoid Toxin-Positive. mSphere 2021, 6, e01255-20. [Google Scholar] [CrossRef]
- Byres, E.; Paton, A.W.; Paton, J.C.; Lofling, J.C.; Smith, D.F.; Wilce, M.C.; Talbot, U.M.; Chong, D.C.; Yu, H.; Huang, S.; et al. Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature 2008, 456, 648–652. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Deng, L.; Stack, G.; Yu, H.; Chen, X.; Naito-Matsui, Y.; Varki, A.; Galán, J.E. Evolution of host adaptation in the Salmonella typhoid toxin. Nat. Microbiol. 2017, 2, 1592–1599. [Google Scholar] [CrossRef]
- Deng, L.; Song, J.; Gao, X.; Wang, J.; Yu, H.; Chen, X.; Varki, N.; Naito-Matsui, Y.; Galán, J.E.; Varki, A. Host adaptation of a bacterial toxin from the human pathogen Salmonella Typhi. Cell 2014, 159, 1290–1299. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Kuziemko, G.M.; Stroh, M.; Stevens, R.C. Cholera toxin binding affinity and specificity for gangliosides determined by surface plasmon resonance. Biochemistry 1996, 35, 6375–6384. [Google Scholar] [CrossRef]
- Holmgren, J.; Lönnroth, I.; Månsson, J.; Svennerholm, L. Interaction of cholera toxin and membrane GM1 ganglioside of small intestine. Proc. Natl. Acad. Sci. USA 1975, 72, 2520–2524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, P.; Klein, M. Single-step purification of pertussis toxin and its subunits by heat-treated fetuin-sepharose affinity chromatography. Biochem. Cell Biol. 1989, 67, 387–391. [Google Scholar] [CrossRef]
- Pande, A.H.; Moe, D.; Jamnadas, M.; Tatulian, S.A.; Teter, K. The pertussis toxin S1 subunit is a thermally unstable protein susceptible to degradation by the 20S proteasome. Biochemistry 2006, 45, 13734–13740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillenius, P.; Jäätmaa, E.; Askelöf, P.; Granström, M.; Tiru, M. The standardization of an assay for pertussis toxin and antitoxin in microplate culture of Chinese hamster ovary cells. J. Biol. Stand. 1985, 13, 61–66. [Google Scholar] [CrossRef]
- Stavric, S.; Speirs, J.I.; Konowalchuk, J.; Jeffrey, D. Stimulation of cyclic AMP secretion in Vero cells by enterotoxins of Escherichia coli and Vibrio cholerae. Infect. Immun. 1978, 21, 514–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feoktistova, M.; Geserick, P.; Leverkus, M. Crystal Violet Assay for Determining Viability of Cultured Cells. Cold Spring Harb. Protoc. 2016, 2016, pdb.prot087379. [Google Scholar] [CrossRef] [PubMed]
- Krueger, K.M.; Mende-Mueller, L.M.; Barbieri, J.T. Protease treatment of pertussis toxin identifies the preferential cleavage of the S1 subunit. J. Biol. Chem. 1991, 266, 8122–8128. [Google Scholar] [CrossRef]
- Gill, D.M.; Rappaport, R.S. Origin of the enzymatically active A1 fragment of cholera toxin. J. Infect. Dis. 1979, 139, 674–680. [Google Scholar] [CrossRef]
- Stein, P.E.; Boodhoo, A.; Armstrong, G.D.; Cockle, S.A.; Klein, M.H.; Read, R.J. The crystal structure of pertussis toxin. Structure 1994, 2, 45–57. [Google Scholar] [CrossRef] [Green Version]
- Carbonetti, N.H.; Mays, R.M.; Artamonova, G.V.; Plaut, R.D.; Worthington, Z.E. Proteolytic cleavage of pertussis toxin S1 subunit is not essential for its activity in mammalian cells. BMC Microbiol. 2005, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- Plaut, R.D.; Scanlon, K.M.; Taylor, M.; Teter, K.; Carbonetti, N.H. Intracellular disassembly and activity of pertussis toxin require interaction with ATP. Pathog. Dis. 2016, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cherubin, P.; Fidler, D.; Quiñones, B.; Teter, K. Bimodal Response to Shiga Toxin 2 Subtypes Results from Relatively Weak Binding to the Target Cell. Infect. Immun. 2019, 87, e00428-19. [Google Scholar] [CrossRef] [PubMed]
- Cherubin, P.; Quiñones, B.; Teter, K. Cellular recovery from exposure to sub-optimal concentrations of AB toxins that inhibit protein synthesis. Sci. Rep. 2018, 8, 2494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glineur, C.; Locht, C. Importance of ADP-ribosylation in the morphological changes of PC12 cells induced by cholera toxin. Infect. Immun. 1994, 62, 4176–4185. [Google Scholar] [CrossRef] [Green Version]
- Fishman, P.H.; Atikkan, E.E. Mechanism of action of cholera toxin: Effect of receptor density and multivalent binding on activation of adenylate cyclase. J. Membr. Biol. 1980, 54, 51–60. [Google Scholar] [CrossRef]
- Maneval, D.R.; Colwell, R.R.; Grays, S.W.J.; Donta, S.T. A tissue culture method for the selection of bacterial enterotoxins. J. Tissue Cult. Methods 1981, 6, 85–90. [Google Scholar] [CrossRef]
- Guerrant, R.L.; Brunton, L.L. Characterization of the Chinese hamster ovary cell assay for the enterotoxins of Vibrio cholerae and Escherichia coli and for specific antisera, and toxoid. J. Infect. Dis. 1977, 135, 720–728. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, R.T.; Yokota, T.; Kuwahara, S. Assay method for Vibrio cholerae and Escherichia coli enterotoxins by automated counting of floating chinese hamster ovary cells in culture medium. J. Clin. Microbiol. 1978, 7, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Terrinoni, M.; Holmgren, J.; Lebens, M.; Larena, M. Proteomic analysis of cholera toxin adjuvant-stimulated human monocytes identifies Thrombospondin-1 and Integrin-β1 as strongly upregulated molecules involved in adjuvant activity. Sci. Rep. 2019, 9, 2812. [Google Scholar] [CrossRef] [Green Version]
- Hewlett, E.L.; Sauer, K.T.; Myers, G.A.; Cowell, J.L.; Guerrant, R.L. Induction of a novel morphological response in Chinese hamster ovary cells by pertussis toxin. Infect. Immun. 1983, 40, 1198–1203. [Google Scholar] [CrossRef] [Green Version]
- Burns, D.L.; Kenimer, J.G.; Manclark, C.R. Role of the A subunit of pertussis toxin in alteration of Chinese hamster ovary cell morphology. Infect. Immun. 1987, 55, 24–28. [Google Scholar] [CrossRef] [Green Version]
- Jobling, M.G. The chromosomal nature of LT-II enterotoxins solved: A lambdoid prophage encodes both LT-II and one of two novel pertussis-toxin-like toxin family members in type II enterotoxigenic Escherichia coli. Pathog. Dis. 2016, 74, ftw001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Creydt, V.P.; Silberstein, C.; Zotta, E.; Ibarra, C. Cytotoxic effect of Shiga toxin-2 holotoxin and its B subunit on human renal tubular epithelial cells. Microbes Infect. 2006, 8, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, Y.; Matano, K.; Asai, T.; Tochikubo, K. Affinity purification of recombinant cholera toxin B subunit oligomer expressed in Bacillus brevis for potential human use as a mucosal adjuvant. FEMS Immunol. Med. Microbiol. 1998, 20, 311–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dugas, B.; Paul-Eugène, N.; Génot, E.; Mencia-Huerta, J.M.; Braquet, P.; Kolb, J.P. Effect of bacterial toxins on human B cell activation. II. Mitogenic activity of the B subunit of cholera toxin. Eur. J. Immunol. 1991, 21, 495–500. [Google Scholar] [CrossRef]
- Smets, L.A.; van Rooy, H. Mitogenic and antimitogenic effects of cholera toxin-mediated cyclic AMP levels in 3T3 cells. J. Cell. Physiol. 1987, 133, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Okada, N.; Kitano, Y.; Ichihara, K. Effects of cholera toxin on proliferation of cultured human keratinocytes in relation to intracellular cyclic AMP levels. J. Invest. Dermatol. 1982, 79, 42–47. [Google Scholar] [CrossRef] [Green Version]
- Allam, M.; Bertrand, R.; Zhang-Sun, G.; Pappas, J.; Viallet, J. Cholera toxin triggers apoptosis in human lung cancer cell lines. Cancer Res. 1997, 57, 2615–2618. [Google Scholar]
- Nashar, T.O.; Williams, N.A.; Hirst, T.R.; Nahar, T.O. Cross-linking of cell surface ganglioside GM1 induces the selective apoptosis of mature CD8+ T lymphocytes. Int. Immunol. 1996, 8, 731–736. [Google Scholar] [CrossRef] [Green Version]
- Aad, G.; Abbott, B.; Abdallah, J.; Abdel Khalek, S.; Abdelalim, A.A.; Abdesselam, A.; Abdinov, O.; Abi, B.; Abolins, M.; Abouzeid, O.S.; et al. Observation of spin correlation in tt events from pp collisions at radicals=7 TeV using the ATLAS detector. Phys. Rev. Lett. 2012, 108, 212001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jobling, M.G.; Holmes, R.K. Identification of motifs in cholera toxin A1 polypeptide that are required for its interaction with human ADP-ribosylation factor 6 in a bacterial two-hybrid system. Proc. Natl. Acad. Sci. USA 2000, 97, 14662–14667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Tinker, J.K.; Davis, C.T.; Arlian, B.M. Purification and characterization of Yersinia enterocolitica and Yersinia pestis LcrV-cholera toxin A(2)/B chimeras. Protein Expr. Purif. 2010, 74, 16–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Overgaard, E.; Morris, B.; Mohammad Mousa, O.; Price, E.; Rodriguez, A.; Cufurovic, L.; Beard, R.S.; Tinker, J.K. Cellular Activity of Salmonella Typhimurium ArtAB Toxin and Its Receptor-Binding Subunit. Toxins 2021, 13, 599. https://doi.org/10.3390/toxins13090599
Overgaard E, Morris B, Mohammad Mousa O, Price E, Rodriguez A, Cufurovic L, Beard RS, Tinker JK. Cellular Activity of Salmonella Typhimurium ArtAB Toxin and Its Receptor-Binding Subunit. Toxins. 2021; 13(9):599. https://doi.org/10.3390/toxins13090599
Chicago/Turabian StyleOvergaard, Elise, Brad Morris, Omid Mohammad Mousa, Emily Price, Adriana Rodriguez, Leyla Cufurovic, Richard S. Beard, and Juliette K. Tinker. 2021. "Cellular Activity of Salmonella Typhimurium ArtAB Toxin and Its Receptor-Binding Subunit" Toxins 13, no. 9: 599. https://doi.org/10.3390/toxins13090599