Membrane-Disrupting Activity of Cobra Cytotoxins Is Determined by Configuration of the N-Terminal Loop
Abstract
:1. Introduction
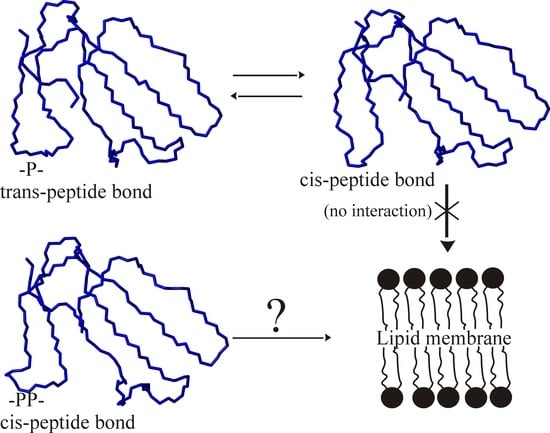
2. Results
3. Discussion
4. Conclusions
5. Material and Methods
5.1. Chemicals
5.2. Isolation of Cytotoxins
5.3. Antibacterial Activity
5.4. Cytotoxic Activity
5.5. Structure Modeling of the Toxins
5.6. Calcein-Loaded Liposome Preparation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Felicio, M.R.; Silva, O.N.; Goncalves, S.; Santos, N.C.; Franco, O.L. Peptides with Dual Antimicrobial and Anticancer Activities. Front. Chem. 2017, 5, 5. [Google Scholar] [CrossRef] [Green Version]
- Hoskin, D.W.; Ramamoorthy, A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta 2008, 1778, 357–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mader, J.S.; Hoskin, D.W. Cationic antimicrobial peptides as novel cytotoxic agents for cancer treatment. Expert. Opin. Investig. Drugs 2006, 15, 933–946. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.; Bhattacharjee, P.; Mishra, R.; Biswas, A.K.; Dasgupta, S.C.; Giri, B. Anticancer potential of animal venoms and toxins. Ind. J. Exp. Biol. 2010, 48, 93–103. [Google Scholar]
- Wang, L.; Dong, C.; Li, X.; Han, W.; Su, X. Anticancer potential of bioactive peptides from animal sources (Review). Oncol. Rep. 2017, 38, 637–651. [Google Scholar] [CrossRef] [Green Version]
- Yacoub, T.; Rima, M.; Karam, M.; Sabatier, J.-M.; Fajloun, Z. Antimicrobials from Venomous Animals: An Overview. Molecules 2020, 25, 2402. [Google Scholar] [CrossRef]
- Raghuraman, H.; Chattopadhyay, A. Melittin: A membrane-active peptide with diverse functions. Biosci. Rep. 2007, 27, 189–223. [Google Scholar] [CrossRef]
- Kuzmenkov, A.I.; Sachkova, M.Y.; Kovalchuk, S.I.; Grishin, E.V.; Vassilevski, A.A. Lachesana tarabaevi, an expert in membrane-active toxins. Biochem. J. 2016, 473, 2495–2506. [Google Scholar] [CrossRef]
- Falcao, C.B.; Radis-Baptista, G. Crotamine and crotalicidin, membrane active peptides from Crotalus durissus terrificus rattlesnake venom, and their structurally-minimized fragments for applications in medicine and biotechnology. Peptides 2020, 126, 170234. [Google Scholar] [CrossRef]
- Nuri, R.; Shprung, T.; Shai, Y. Defensive remodeling: How bacterial surface properties and biofilm formation promote resistance to antimicrobial peptides. Biochim. Biophys. Acta 2015, 1848, 3089–3100. [Google Scholar] [CrossRef] [Green Version]
- Deslouches, B.; Di, Y.P. Antimicrobial peptides with selective antitumor mechanisms: Prospect for anticancer applications. Oncotarget 2017, 8, 46635–46651. [Google Scholar] [CrossRef]
- Lin, L.; Chi, J.; Yan, Y.; Luo, R.; Feng, X.; Zheng, Y.; Xian, D.; Li, X.; Quan, G.; Liu, D.; et al. Membrane-disruptive peptides/peptidomimetics-based therapeutics: Promising systems to combat bacteria and cancer in the drug-resistant era. Acta Pharm. Sin. B 2021, 11, 2609–2644. [Google Scholar] [CrossRef]
- Bernheimer, A.W.; Rudy, B. Interactions between membranes and cytolytic peptides. Biochim. Biophys. Acta 1986, 864, 123–141. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Vassilevski, A.A.; Kozlov, S.A.; Feofanov, A.V.; Grishin, E.V.; Efremov, R.G. Latarcins: Versatile spider venom peptides. Cell Mol. Life Sci. 2015, 72, 4501–4522. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Efremov, R.G. The role of hydrophobic /hydrophilic balance in the activity of structurally flexible vs. rigid cytolytic polypeptides and analogs developed on their basis. Expert Rev. Proteom. 2018, 15, 873–886. [Google Scholar] [CrossRef]
- Kumar, T.K.; Jayaraman, G.; Lee, C.S.; Arunkumar, A.I.; Sivaraman, T.; Samuel, D.; Yu, C. Snake venom cardiotoxins-structure, dynamics, function and folding. J. Biomol. Struct. Dyn. 1997, 15, 431–463. [Google Scholar] [CrossRef]
- Kini, R.M.; Doley, R. Structure, function and evolution of three-finger toxins: Mini proteins with multiple targets. Toxicon 2010, 56, 855–867. [Google Scholar] [CrossRef]
- Gasanov, S.E.; Dagda, R.K.; Rael, E.D. Snake Venom Cytotoxins, Phospholipase A2s, and Zn-dependent Metalloproteinases: Mechanisms of Action and Pharmacological Relevance. J. Clin. Toxicol. 2014, 4, 1000181. [Google Scholar] [CrossRef] [Green Version]
- Dubovskii, P.V.; Utkin, Y.N. Antiproliferative activity of cobra venom cytotoxins. Curr. Top. Med. Chem. 2015, 15, 638–648. [Google Scholar] [CrossRef]
- Konshina, A.G.; Dubovskii, P.V.; Efremov, R.G. Structure and dynamics of cardiotoxins. Curr. Prot. Pept. Sci. 2012, 13, 570–584. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Utkin, Y.N. Cobra cytotoxins: Structural organization and antibacterial activity. Acta Nat. 2014, 6, 11–18. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Konshina, A.G.; Efremov, R.G. Cobra cardiotoxins: Membrane interactions and pharmacological potential. Curr. Med. Chem. 2014, 21, 270–287. [Google Scholar] [CrossRef] [PubMed]
- Feofanov, A.V.; Sharonov, G.V.; Astapova, M.V.; Rodionov, D.I.; Utkin, Y.N.; Arseniev, A.S. Cancer cell injury by cytotoxins from cobra venom is mediated through lysosomal damage. Biochem. J. 2005, 390, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Jain, D.; Kumar, S. Snake Venom: A Potent Anticancer Agent. Asian Pac. J. Cancer Prev. 2012, 13, 4855–4860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebrahim, K.; Shirazi, F.H.; Mirakabadi, A.Z.; Vatanpour, H. Cobra venom cytotoxins; apoptotic or necrotic agents? Toxicon 2015, 108, 134–140. [Google Scholar] [CrossRef]
- Abidin, Z.; Asnawi, S.; Lee, Y.Q.; Othman, I.; Naidu, R. Malaysian Cobra Venom: A Potential Source of Anti-Cancer Therapeutic Agents. Toxins 2019, 11, 75. [Google Scholar] [CrossRef] [Green Version]
- Efremov, R.G.; Volynsky, P.E.; Nolde, D.E.; Dubovskii, P.V.; Arseniev, A.S. Interaction of cardiotoxins with membranes: A molecular modeling study. Biophys. J. 2002, 83, 144–153. [Google Scholar] [CrossRef] [Green Version]
- Dementieva, D.V.; Bocharov, E.V.; Arseniev, A.S. Two forms of cytotoxin II (cardiotoxin) from Naja naja oxiana in aqueous solution: Spatial structures with tightly bound water molecules. Eur. J. Biochem. 1999, 263, 152–162. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Dubinnyi, M.A.; Konshina, A.G.; Kazakova, E.D.; Sorokoumova, G.M.; Ilyasova, T.M.; Shulepko, M.A.; Chertkova, R.V.; Lyukmanova, E.N.; Dolgikh, D.A.; et al. Structural and Dynamic “Portraits” of Recombinant and Native Cytotoxin I from Naja oxiana: How Close Are They? Biochemistry 2017, 56, 4468–4477. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Dubinny, M.A.; Volynsky, P.E.; Pustovalova, Y.E.; Konshina, A.G.; Utkin, Y.N.; Efremov, R.G.; Arseniev, A.S. Impact of membrane partitioning on the spatial structure of an S-type cobra cytotoxin. J. Biomol. Struct. Dyn. 2018, 36, 3463–3478. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Dementieva, D.V.; Bocharov, E.V.; Utkin, Y.N.; Arseniev, A.S. Membrane binding motif of the P-type cardiotoxin. J. Mol. Biol. 2001, 305, 137–149. [Google Scholar] [CrossRef]
- Konshina, A.G.; Dubovskii, P.V.; Efremov, R.G. Stepwise Insertion of Cobra Cardiotoxin CT2 into a Lipid Bilayer Occurs as an Interplay of Protein and Membrane “Dynamic Molecular Portraits”. J. Chem. Inf. Model. 2021, 61, 385–399. [Google Scholar] [CrossRef]
- Dauplais, M.; Neumann, J.M.; Pinkasfeld, S.; Menez, A.; Roumestand, C. An NMR study of the interaction of cardiotoxin gamma from Naja nigricollis with perdeuterated dodecylphosphocholine micelles. Eur. J. Biochem. 1995, 230, 213–220. [Google Scholar] [CrossRef]
- Chen, T.-S.; Chung, F.-Y.; Tjong, S.-C.; Goh, K.-S.; Huang, W.-N.; Chien, K.-Y.; Wu, P.-L.; Lin, H.-C.; Chen, C.-J.; Wu, W.-G. Structural difference between group I and group II cobra cardiotoxins: X-ray, NMR, and CD analysis of the effect of cis-proline conformation on three-fingered toxins. Biochemistry 2005, 44, 7414–7426. [Google Scholar] [CrossRef]
- Utkin, Y.N.; Kuch, U.; Kasheverov, I.E.; Lebedev, D.S.; Cederlund, E.; Molles, B.E.; Polyak, I.; Ivanov, I.A.; Prokopev, N.A.; Ziganshin, R.H.; et al. Novel long-chain neurotoxins from Bungarus candidus distinguish the two binding sites in muscle-type nicotinic acetylcholine receptors. Biochem. J. 2019, 476, 1285–1302. [Google Scholar] [CrossRef]
- Suzuki, M.; Itoh, T.; Anuruddhe, B.M.; Bandaranayake, I.K.; Ranasinghe, J.G.S.; Athauda, S.B.; Moriyama, A. Molecular diversity in venom proteins of the Russell’s viper (Daboia russellii russellii) and the Indian cobra (Naja naja) in Sri Lanka. Biomed. Res. 2010, 31, 71–81. [Google Scholar] [CrossRef] [Green Version]
- Dubovskii, P.V.; Dubova, K.M.; Bourenkov, G.; Starkov, V.G.; Konshina, A.G.; Efremov, R.G.; Utkin, Y.N.; Samygina, V.R. Variability in the Spatial Structure of the Central Loop in Cobra Cytotoxins Revealed by X-ray Analysis and Molecular Modeling. Toxins 2022, 14, 149. [Google Scholar] [CrossRef]
- Dubinnyi, M.A.; Dubovskii, P.V.; Starkov, V.A.; Utkin, Y.N. The omega-loop of cobra cytotoxins tolerates multiple amino acid substitutions. Biochem. Biophys. Res. Commun. 2021, 558, 141–146. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Vorontsova, O.V.; Utkin, Y.N.; Arseniev, A.S.; Efremov, R.G.; Feofanov, A.V. Cobra cytotoxins: Determinants of antibacterial activity. Mendeleev Commun. 2015, 21, 70–71. [Google Scholar] [CrossRef]
- Weise, K.H.; Carlsson, F.H.; Joubert, F.J.; Strydom, D.J. Snake venom toxins. The purification of toxins VII1 and VII2, two cytotoxin homologues from banded Egyptian cobra (Naja haje annulifera) venom, and the complete amino acid sequence of toxin VII1. Hoppe Seylers Z Physiol. Chem. 1973, 354, 1317–1326. [Google Scholar] [CrossRef]
- Suryamohan, K.; Krishnankutty, S.P.; Guillory, J.; Jevit, M.; Schroder, M.S.; Wu, M.; Kuriakose, B.; Mathew, O.K.; Perumal, R.C.; Koludarov, I.; et al. The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat. Genet. 2020, 52, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Sivaraman, T.; Kumar, T.K.; Yang, P.W.; Yu, C. Cardiotoxin-like basic protein (CLBP) from Naja naja atra is not a cardiotoxin. Toxicon 1997, 35, 1367–1371. [Google Scholar] [CrossRef] [PubMed]
- Dubovskii, P.V.; Ignatova, A.A.; Feofanov, A.V.; Utkin, Y.N.; Efremov, R.G. Antibacterial activity of cardiotoxin-like basic polypeptide from cobra venom. Bioorg. Med. Chem. Lett. 2020, 30, 126890. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-C.; Lin, C.-C.; Wang, C.-H.; Wu, P.-L.; Huang, H.-W.; Chang, C.-I.; Wu, W.-G. Endocytotic routes of cobra cardiotoxins depend on spatial distribution of positively charged and hydrophobic domains to target distinct types of sulfated glycoconjugates on cell surface. J. Biol. Chem. 2014, 289, 20170–20181. [Google Scholar] [CrossRef] [Green Version]
- Bordoli, L.; Kiefer, F.; Arnold, K.; Benkert, P.; Battey, J.; Schwede, T. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 2009, 4, 1–13. [Google Scholar] [CrossRef]







| Toxin Name | Uniprot Codes of the Identified Toxins | Molecular Weight, Da 1 | Isoelectric Point | Electrical Charge | P/S-Type | Antibacterial (B. subtilis) Effect, MIC, µM | Cytotoxic Effect (Against A549 cells), LD50, µM |
|---|---|---|---|---|---|---|---|
| Nn17-3 | P01440 | 6755.2 | 9.36 | 9 | P | 0.8 | 7.4 ± 0.1 |
| Nn18-3 | - | 6782.5 | 9.36 | 9 2 | P 2 | 0.8 | 5.0 ± 0.5 |
| Nn16-1 | P01445 | 6736.5 | 9.38 | 9 | S | 3.1 | 5.6 ± 1.5 |
| Nn15-1 | P01447-1 | 6783.5 | 9.24 | 8 | S | 6.3 | 7.9 ± 0.4 |
| Nn14-1 | P86382-1 | 6783.7 | 9.11 | 7 | S | 50 | 17.4 ± 0.8 |
| Nh1 3 | P01455 | 6688.1 | 9.15 | 8 | S | >50 (80 4) | 132 ± 9 5 |
| Nh2 3 | P01462 | 6850.3 | 8.99 | 7 | P | >50 (40 4) | 116 ± 6 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dubovskii, P.V.; Ignatova, A.A.; Alekseeva, A.S.; Starkov, V.G.; Boldyrev, I.A.; Feofanov, A.V.; Utkin, Y.N. Membrane-Disrupting Activity of Cobra Cytotoxins Is Determined by Configuration of the N-Terminal Loop. Toxins 2023, 15, 6. https://doi.org/10.3390/toxins15010006
Dubovskii PV, Ignatova AA, Alekseeva AS, Starkov VG, Boldyrev IA, Feofanov AV, Utkin YN. Membrane-Disrupting Activity of Cobra Cytotoxins Is Determined by Configuration of the N-Terminal Loop. Toxins. 2023; 15(1):6. https://doi.org/10.3390/toxins15010006
Chicago/Turabian StyleDubovskii, Peter V., Anastasia A. Ignatova, Anna S. Alekseeva, Vladislav G. Starkov, Ivan A. Boldyrev, Alexey V. Feofanov, and Yuri N. Utkin. 2023. "Membrane-Disrupting Activity of Cobra Cytotoxins Is Determined by Configuration of the N-Terminal Loop" Toxins 15, no. 1: 6. https://doi.org/10.3390/toxins15010006