Modulation of Autophagy and Cell Death by Bacterial Outer-Membrane Vesicles
Abstract
:1. Diversity, Biogenesis and Functions of Bacteria Extracellular Vesicles
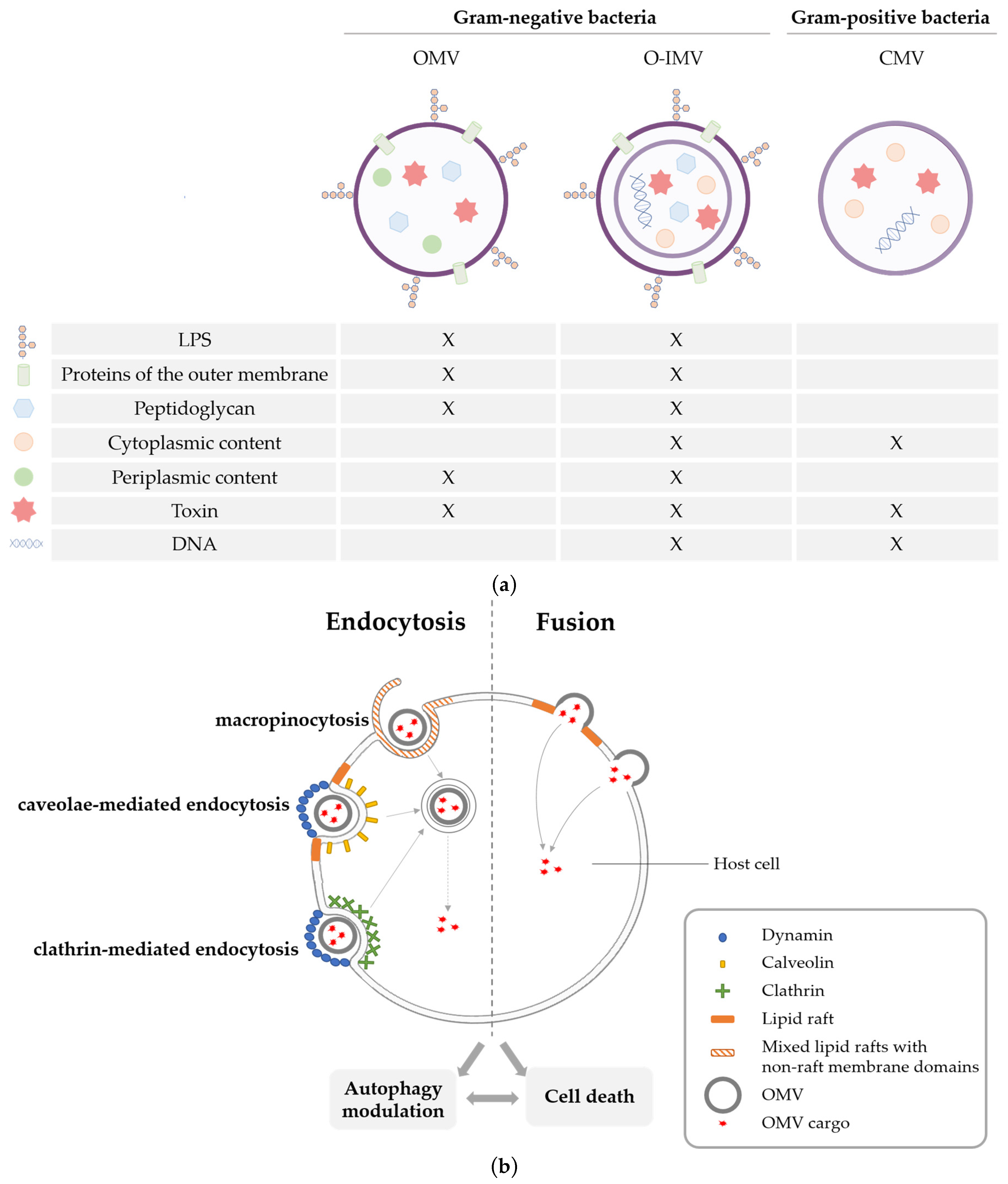
2. Cell Death Pathways Triggered by OMVs
2.1. OMVs Induced Pyroptosis
2.2. OMVs Induced Apoptosis
3. Modulation of Autophagy by OMVs and Consequences on Cell Fate
3.1. Autophagy Induction as an Antimicrobial Mechanism
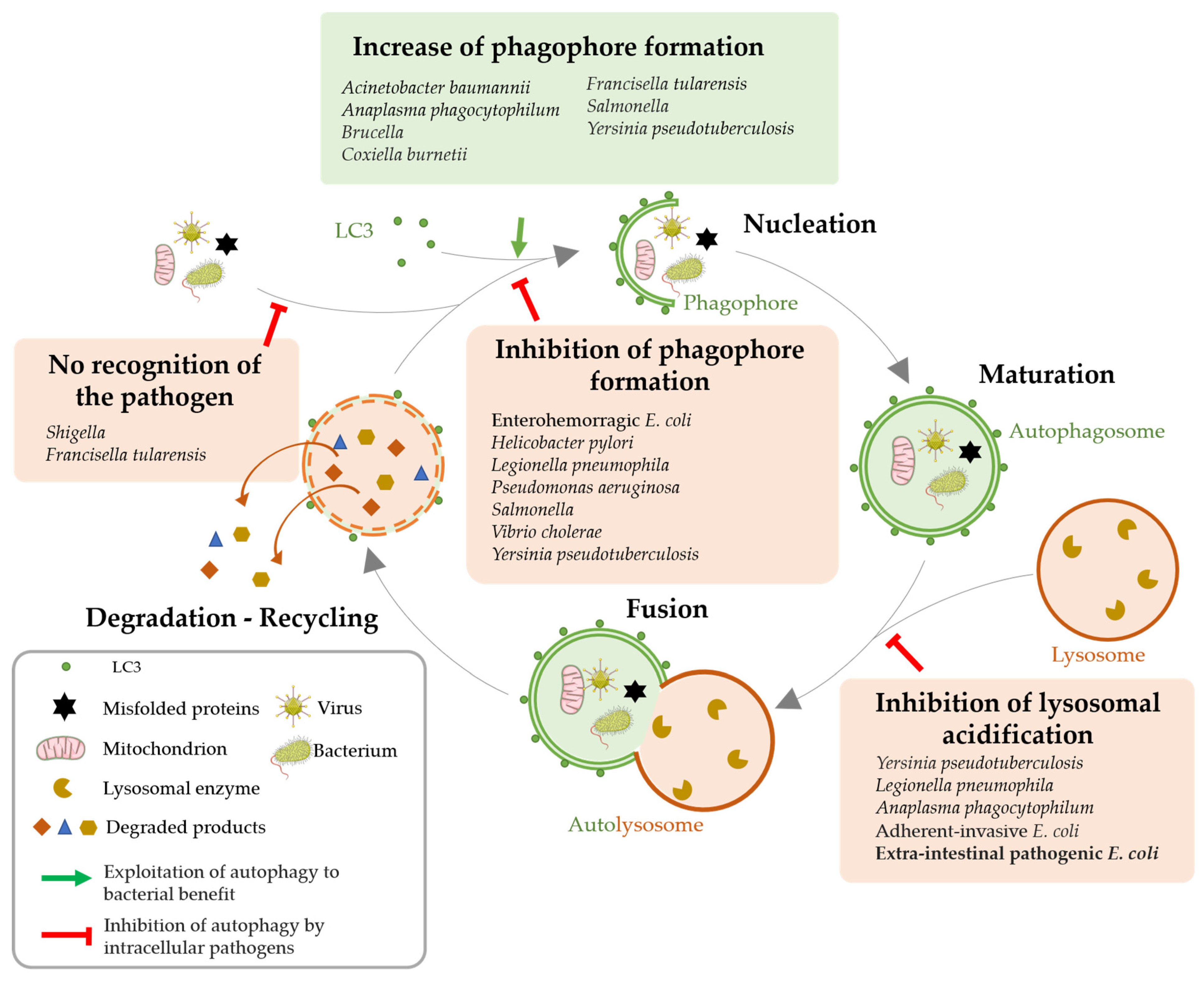
3.2. Autophagy Impairment for Bacterial Benefit
3.3. Consequences of Autophagy Modulation by Pathogenic Bacteria
4. Concluding Remarks and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gill, S.; Catchpole, R.; Forterre, P. Extracellular Membrane Vesicles in the Three Domains of Life and Beyond. FEMS Microbiol. Rev. 2019, 43, 273–303. [Google Scholar] [CrossRef] [PubMed]
- Briaud, P.; Carroll, R.K. Extracellular Vesicle Biogenesis and Functions in Gram-Positive Bacteria. Infect. Immun. 2020, 88, e00433-20. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Lin, H. Characterization and Function of Membrane Vesicles in Gram-Positive Bacteria. Appl. Microbiol. Biotechnol. 2021, 105, 1795–1801. [Google Scholar] [CrossRef] [PubMed]
- Palacios, A.; Gupta, S.; Rodriguez, G.M.; Prados-Rosales, R. Extracellular Vesicles in the Context of Mycobacterium Tuberculosis Infection. Mol. Immunol. 2021, 133, 175–181. [Google Scholar] [CrossRef]
- Pérez-Cruz, C.; Carriòn, O.; Delgado, L.; Martinez, G.; Lòpez-Iglesias, C.; Mercade, E. New Type of Outer Membrane Vesicle Produced by the Gram-Negative Bacterium Shewanella Vesiculosa M7T: Implications for DNA Content. Appl. Environ. Microbiol. 2013, 79, 1874–1881. [Google Scholar] [CrossRef]
- Turnbull, L.; Toyofuku, M.; Hynen, A.L.; Kurosawa, M.; Pessi, G.; Petty, N.K.; Osvath, S.R.; Cárcamo-Oyarce, G.; Gloag, E.S.; Shimoni, R.; et al. Explosive Cell Lysis as a Mechanism for the Biogenesis of Bacterial Membrane Vesicles and Biofilms. Nat. Commun. 2016, 7, 11220. [Google Scholar] [CrossRef]
- Tashiro, Y. Bacterial Membrane Vesicles with Multiple Lipid Bilayers: Vesicles Harboring Organelle-like Structures. Biosci. Biotechnol. Biochem. 2022, 86, 967–973. [Google Scholar] [CrossRef]
- Toyofuku, M.; Schild, S.; Kaparakis-Liaskos, M.; Eberl, L. Composition and Functions of Bacterial Membrane Vesicles. Nat. Rev. Microbiol. 2023, 21, 415–430. [Google Scholar] [CrossRef]
- O’Donoghue, E.J.; Krachler, A.M. Mechanisms of Outer Membrane Vesicle Entry into Host Cells. Cell. Microbiol. 2016, 18, 1508–1517. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Kuehn, M.J. Outer-Membrane Vesicles from Gram-Negative Bacteria: Biogenesis and Functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef]
- Giordano, N.P.; Cian, M.B.; Dalebroux, Z.D. Outer Membrane Lipid Secretion and the Innate Immune Response to Gram-Negative Bacteria. Infect. Immun. 2020, 88, e00920-19. [Google Scholar] [CrossRef]
- Tashiro, Y.; Inagaki, A.; Shimizu, M.; Ichikawa, S.; Takaya, N.; Nakajima-Kambe, T.; Uchiyama, H.; Nomura, N. Characterization of Phospholipids in Membrane Vesicles Derived from Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 2011, 75, 605–607. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C.; Kulp, A.; Kuehn, M.J. Modulation of Bacterial Outer Membrane Vesicle Production by Envelope Structure and Content. BMC Microbiol. 2014, 14, 324. [Google Scholar] [CrossRef] [PubMed]
- McBroom, A.J.; Kuehn, M.J. Release of Outer Membrane Vesicles by Gram-Negative Bacteria Is a Novel Envelope Stress Response. Mol. Microbiol. 2007, 63, 545–558. [Google Scholar] [CrossRef]
- Tashiro, Y.; Ichikawa, S.; Nakajima-Kambe, T.; Uchiyama, H.; Nomura, N. Pseudomonas Quinolone Signal Affects Membrane Vesicle Production in Not Only Gram-Negative but Also Gram-Positive Bacteria. Microbes Environ. 2010, 25, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Murase, K.; Martin, P.; Porcheron, G.; Houle, S.; Helloin, E.; Pénary, M.; Nougayrède, J.-P.; Dozois, C.M.; Hayashi, T.; Oswald, E. HlyF Produced by Extraintestinal Pathogenic Escherichia coli Is a Virulence Factor That Regulates Outer Membrane Vesicle Biogenesis. J. Infect. Dis. 2016, 213, 856–865. [Google Scholar] [CrossRef]
- Zhuge, X.; Sun, Y.; Xue, F.; Tang, F.; Ren, J.; Li, D.; Wang, J.; Jiang, M.; Dai, J. A Novel PhoP/PhoQ Regulation Pathway Modulates the Survival of Extraintestinal Pathogenic Escherichia coli in Macrophages. Front. Immunol. 2018, 9, 788. [Google Scholar] [CrossRef]
- Guerrero-Mandujano, A.; Hernández-Cortez, C.; Ibarra, J.A.; Castro-Escarpulli, G. The Outer Membrane Vesicles: Secretion System Type Zero. Traffic 2017, 18, 425–432. [Google Scholar] [CrossRef]
- Rueter, C.; Bielaszewska, M. Secretion and Delivery of Intestinal Pathogenic Escherichia coli Virulence Factors via Outer Membrane Vesicles. Front. Cell. Infect. Microbiol. 2020, 10, 91. [Google Scholar] [CrossRef]
- Bielaszewska, M.; Rüter, C.; Bauwens, A.; Greune, L.; Jarosch, K.-A.; Steil, D.; Zhang, W.; He, X.; Lloubes, R.; Fruth, A.; et al. Host Cell Interactions of Outer Membrane Vesicle-Associated Virulence Factors of Enterohemorrhagic Escherichia coli O157: Intracellular Delivery, Trafficking and Mechanisms of Cell Injury. PLoS Pathog. 2017, 13, e1006159. [Google Scholar] [CrossRef]
- Bomberger, J.M.; MacEachran, D.P.; Coutermarsh, B.A.; Ye, S.; O’Toole, G.A.; Stanton, B.A. Long-Distance Delivery of Bacterial Virulence Factors by Pseudomonas aeruginosa Outer Membrane Vesicles. PLoS Pathog. 2009, 5, e1000382. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, D.; Chaudhuri, K. Association of Cholera Toxin with Vibrio Cholerae Outer Membrane Vesicles Which Are Internalized by Human Intestinal Epithelial Cells. FEBS Lett. 2011, 585, 1357–1362. [Google Scholar] [CrossRef] [PubMed]
- Turner, L.; Bitto, N.J.; Steer, D.L.; Lo, C.; D’Costa, K.; Ramm, G.; Shambrook, M.; Hill, A.F.; Ferrero, R.L.; Kaparakis-Liaskos, M. Helicobacter pylori Outer Membrane Vesicle Size Determines Their Mechanisms of Host Cell Entry and Protein Content. Front. Immunol. 2018, 9, 1466. [Google Scholar] [CrossRef] [PubMed]
- O’Donoghue, E.J.; Sirisaengtaksin, N.; Browning, D.F.; Bielska, E.; Hadis, M.; Fernandez-Trillo, F.; Alderwick, L.; Jabbari, S.; Krachler, A.M. Lipopolysaccharide Structure Impacts the Entry Kinetics of Bacterial Outer Membrane Vesicles into Host Cells. PLoS Pathog. 2017, 13, e1006760. [Google Scholar] [CrossRef]
- Zingl, F.G.; Thapa, H.B.; Scharf, M.; Kohl, P.; Müller, A.M.; Schild, S. Outer Membrane Vesicles of Vibrio Cholerae Protect and Deliver Active Cholera Toxin to Host Cells via Porin-Dependent Uptake. mBio 2021, 12, e00534-21. [Google Scholar] [CrossRef]
- Kuehn, M.J.; Kesty, N.C. Bacterial Outer Membrane Vesicles and the Host–Pathogen Interaction. Genes Dev. 2005, 19, 2645–2655. [Google Scholar] [CrossRef]
- Jin, J.S.; Kwon, S.-O.; Moon, D.C.; Gurung, M.; Lee, J.H.; Kim, S.I.; Lee, J.C. Acinetobacter Baumannii Secretes Cytotoxic Outer Membrane Protein A via Outer Membrane Vesicles. PLoS ONE 2011, 6, e17027. [Google Scholar] [CrossRef]
- Krsek, D.; Yara, D.A.; Hrbáčková, H.; Daniel, O.; Mančíková, A.; Schüller, S.; Bielaszewska, M. Translocation of Outer Membrane Vesicles from Enterohemorrhagic Escherichia coli O157 across the Intestinal Epithelial Barrier. Front. Microbiol. 2023, 14, 1198945. [Google Scholar] [CrossRef]
- Bittel, M.; Reichert, P.; Sarfati, I.; Dressel, A.; Leikam, S.; Uderhardt, S.; Stolzer, I.; Phu, T.A.; Ng, M.; Vu, N.K.; et al. Visualizing Transfer of Microbial Biomolecules by Outer Membrane Vesicles in Microbe-host-communication in Vivo. J. Extracell. Vesicles 2021, 10, e12159. [Google Scholar] [CrossRef]
- Tulkens, J.; De Wever, O.; Hendrix, A. Analyzing Bacterial Extracellular Vesicles in Human Body Fluids by Orthogonal Biophysical Separation and Biochemical Characterization. Nat. Protoc. 2020, 15, 40–67. [Google Scholar] [CrossRef]
- Hilbi, H.; Moss, J.E.; Hersh, D.; Chen, Y.; Arondel, J.; Banerjee, S.; Flavell, R.A.; Yuan, J.; Sansonetti, P.J.; Zychlinsky, A. Shigella-Induced Apoptosis Is Dependent on Caspase-1 Which Binds to IpaB. J. Biol. Chem. 1998, 273, 32895–32900. [Google Scholar] [CrossRef]
- Cookson, B.T.; Brennan, M.A. Pro-Inflammatory Programmed Cell Death. Trends Microbiol. 2001, 9, 113–114. [Google Scholar] [CrossRef] [PubMed]
- Kist, M.; Vucic, D. Cell Death Pathways: Intricate Connections and Disease Implications. EMBO J. 2021, 40, e106700. [Google Scholar] [CrossRef] [PubMed]
- Garib, F.Y.; Rizopulu, A.P.; Kuchmiy, A.A.; Garib, V.F. Inactivation of Inflammasomes by Pathogens Regulates Inflammation. Biochem. Biokhimiia 2016, 81, 1326–1339. [Google Scholar] [CrossRef]
- Yang, J.; Hwang, I.; Lee, E.; Shin, S.J.; Lee, E.-J.; Rhee, J.H.; Yu, J.-W. Bacterial Outer Membrane Vesicle-Mediated Cytosolic Delivery of Flagellin Triggers Host NLRC4 Canonical Inflammasome Signaling. Front. Immunol. 2020, 11, 581165. [Google Scholar] [CrossRef] [PubMed]
- Vanaja, S.K.; Russo, A.J.; Behl, B.; Banerjee, I.; Yankova, M.; Deshmukh, S.D.; Rathinam, V.A.K. Bacterial Outer Membrane Vesicles Mediate Cytosolic Localization of LPS and Caspase-11 Activation. Cell 2016, 165, 1106–1119. [Google Scholar] [CrossRef]
- Santos, J.C.; Dick, M.S.; Lagrange, B.; Degrandi, D.; Pfeffer, K.; Yamamoto, M.; Meunier, E.; Pelczar, P.; Henry, T.; Broz, P. LPS Targets Host Guanylate-binding Proteins to the Bacterial Outer Membrane for Non-canonical Inflammasome Activation. EMBO J. 2018, 37, e98089. [Google Scholar] [CrossRef]
- Elizagaray, M.L.; Gomes, M.T.R.; Guimaraes, E.S.; Rumbo, M.; Hozbor, D.F.; Oliveira, S.C.; Moreno, G. Canonical and Non-Canonical Inflammasome Activation by Outer Membrane Vesicles Derived From Bordetella Pertussis. Front. Immunol. 2020, 11, 1879. [Google Scholar] [CrossRef]
- Fleetwood, A.J.; Lee, M.K.S.; Singleton, W.; Achuthan, A.; Lee, M.-C.; O’Brien-Simpson, N.M.; Cook, A.D.; Murphy, A.J.; Dashper, S.G.; Reynolds, E.C.; et al. Metabolic Remodeling, Inflammasome Activation, and Pyroptosis in Macrophages Stimulated by Porphyromonas Gingivalis and Its Outer Membrane Vesicles. Front. Cell. Infect. Microbiol. 2017, 7, 351. [Google Scholar] [CrossRef]
- Kaparakis, M.; Turnbull, L.; Carneiro, L.; Firth, S.; Coleman, H.A.; Parkington, H.C.; Le Bourhis, L.; Karrar, A.; Viala, J.; Mak, J.; et al. Bacterial Membrane Vesicles Deliver Peptidoglycan to NOD1 in Epithelial Cells. Cell. Microbiol. 2010, 12, 372–385. [Google Scholar] [CrossRef]
- Irving, A.T.; Mimuro, H.; Kufer, T.A.; Lo, C.; Wheeler, R.; Turner, L.J.; Thomas, B.J.; Malosse, C.; Gantier, M.P.; Casillas, L.N.; et al. The Immune Receptor NOD1 and Kinase RIP2 Interact with Bacterial Peptidoglycan on Early Endosomes to Promote Autophagy and Inflammatory Signaling. Cell Host Microbe 2014, 15, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.F.R.; Wyllie, A.H.; Currie, A.R. Apoptosis: A Basic Biological Phenomenon with Wideranging Implications in Tissue Kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef] [PubMed]
- An, Z.; Huang, X.; Zheng, C.; Ding, W. Acinetobacter Baumannii Outer Membrane Protein A Induces HeLa Cell Autophagy via MAPK/JNK Signaling Pathway. Int. J. Med. Microbiol. 2019, 309, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Tiku, V.; Kofoed, E.M.; Yan, D.; Kang, J.; Xu, M.; Reichelt, M.; Dikic, I.; Tan, M.-W. Outer Membrane Vesicles Containing OmpA Induce Mitochondrial Fragmentation to Promote Pathogenesis of Acinetobacter Baumannii. Sci. Rep. 2021, 11, 618. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.H.; Lee, E.Y.; Lee, Y.C.; Park, T.I.; Kim, H.J.; Hyun, S.H.; Kim, S.A.; Lee, S.-K.; Lee, J.C. Outer Membrane Protein 38 of Acinetobacter Baumannii Localizes to the Mitochondria and Induces Apoptosis of Epithelial Cells. Cell. Microbiol. 2005, 7, 1127–1138. [Google Scholar] [CrossRef]
- Kesavan, D.; Vasudevan, A.; Wu, L.; Chen, J.; Su, Z.; Wang, S.; Xu, H. Integrative Analysis of Outer Membrane Vesicles Proteomics and Whole-Cell Transcriptome Analysis of Eravacycline Induced Acinetobacter Baumannii Strains. BMC Microbiol. 2020, 20, 31. [Google Scholar] [CrossRef]
- Deo, P.; Chow, S.H.; Hay, I.D.; Kleifeld, O.; Costin, A.; Elgass, K.D.; Jiang, J.-H.; Ramm, G.; Gabriel, K.; Dougan, G.; et al. Outer Membrane Vesicles from Neisseria Gonorrhoeae Target PorB to Mitochondria and Induce Apoptosis. PLoS Pathog. 2018, 14, e1006945. [Google Scholar] [CrossRef]
- Bielaszewska, M.; Rüter, C.; Kunsmann, L.; Greune, L.; Bauwens, A.; Zhang, W.; Kuczius, T.; Kim, K.S.; Mellmann, A.; Schmidt, M.A.; et al. Enterohemorrhagic Escherichia coli Hemolysin Employs Outer Membrane Vesicles to Target Mitochondria and Cause Endothelial and Epithelial Apoptosis. PLoS Pathog. 2013, 9, e1003797. [Google Scholar] [CrossRef]
- Deo, P.; Chow, S.H.; Han, M.-L.; Speir, M.; Huang, C.; Schittenhelm, R.B.; Dhital, S.; Emery, J.; Li, J.; Kile, B.T.; et al. Mitochondrial Dysfunction Caused by Outer Membrane Vesicles from Gram-Negative Bacteria Activates Intrinsic Apoptosis and Inflammation. Nat. Microbiol. 2020, 5, 1418–1427. [Google Scholar] [CrossRef]
- Sunshine, H.; Iruela-Arispe, M.L. Membrane Lipids and Cell Signaling. Curr. Opin. Lipidol. 2017, 28, 408–413. [Google Scholar] [CrossRef]
- Arstila, A.U.; Trump, B.F. Studies on Cellular Autophagocytosis. The Formation of Autophagic Vacuoles in the Liver after Glucagon Administration. Am. J. Pathol. 1968, 53, 687–733. [Google Scholar]
- Gomes, L.C.; Dikic, I. Autophagy in Antimicrobial Immunity. Mol. Cell 2014, 54, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Deretic, V.; Levine, B. Autophagy, Immunity, and Microbial Adaptations. Cell Host Microbe 2009, 5, 527–549. [Google Scholar] [CrossRef]
- Vargas, J.N.S.; Hamasaki, M.; Kawabata, T.; Youle, R.J.; Yoshimori, T. The Mechanisms and Roles of Selective Autophagy in Mammals. Nat. Rev. Mol. Cell Biol. 2023, 24, 167–185. [Google Scholar] [CrossRef]
- Kimmey, J.M.; Stallings, C.L. Bacterial Pathogens versus Autophagy: Implications for Therapeutic Interventions. Trends Mol. Med. 2016, 22, 1060–1076. [Google Scholar] [CrossRef]
- Reggio, A.; Buonomo, V.; Grumati, P. Eating the Unknown: Xenophagy and ER-Phagy Are Cytoprotective Defenses against Pathogens. Exp. Cell Res. 2020, 396, 112276. [Google Scholar] [CrossRef]
- Elluri, S.; Enow, C.; Vdovikova, S.; Rompikuntal, P.K.; Dongre, M.; Carlsson, S.; Pal, A.; Uhlin, B.E.; Wai, S.N. Outer Membrane Vesicles Mediate Transport of Biologically Active Vibrio Cholerae Cytolysin (VCC) from V. Cholerae Strains. PLoS ONE 2014, 9, e106731. [Google Scholar] [CrossRef]
- Losier, T.T.; Akuma, M.; McKee-Muir, O.C.; LeBlond, N.D.; Suk, Y.; Alsaadi, R.M.; Guo, Z.; Reshke, R.; Sad, S.; Campbell-Valois, F.-X.; et al. AMPK Promotes Xenophagy through Priming of Autophagic Kinases upon Detection of Bacterial Outer Membrane Vesicles. Cell Rep. 2019, 26, 2150–2165.e5. [Google Scholar] [CrossRef] [PubMed]
- Losier, T.T.; Russell, R.C. Bacterial Outer Membrane Vesicles Trigger Pre-Activation of a Xenophagic Response via AMPK. Autophagy 2019, 15, 1489–1491. [Google Scholar] [CrossRef] [PubMed]
- Escoll, P.; Rolando, M.; Buchrieser, C. Modulation of Host Autophagy during Bacterial Infection: Sabotaging Host Munitions for Pathogen Nutrition. Front. Immunol. 2016, 7, 81. [Google Scholar] [CrossRef] [PubMed]
- Randow, F.; Youle, R.J. Self and Nonself: How Autophagy Targets Mitochondria and Bacteria. Cell Host Microbe 2014, 15, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.H.; Song, H.K. A Structural View of Xenophagy, a Battle between Host and Microbes. Mol. Cells 2018, 41, 27–34. [Google Scholar] [CrossRef]
- Deretic, V.; Saitoh, T.; Akira, S. Autophagy in Infection, Inflammation and Immunity. Nat. Rev. Immunol. 2013, 13, 722–737. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, X.; Wang, Y.; Wang, H.; Huang, C.; Li, J. Endoplasmic Reticulum Stress-Induced Hepatic Stellate Cell Apoptosis through Calcium-Mediated JNK/P38 MAPK and Calpain/Caspase-12 Pathways. Mol. Cell. Biochem. 2014, 394, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Yoshimori, T.; Suzuki, T.; Sagara, H.; Mizushima, N.; Sasakawa, C. Escape of Intracellular Shigella from Autophagy. Science 2005, 307, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Rao, L.; De La Rosa, I.; Xu, Y.; Sha, Y.; Bhattacharya, A.; Holtzman, M.J.; Gilbert, B.E.; Eissa, N.T. Pseudomonas aeruginosa Survives in Epithelia by ExoS-mediated Inhibition of Autophagy and MTOR. EMBO Rep. 2021, 22, e50613. [Google Scholar] [CrossRef] [PubMed]
- Valencia Lopez, M.J.; Schimmeck, H.; Gropengießer, J.; Middendorf, L.; Quitmann, M.; Schneider, C.; Holstermann, B.; Wacker, R.; Heussler, V.; Reimer, R.; et al. Activation of the Macroautophagy Pathway by Yersinia enterocolitica Promotes Intracellular Multiplication and Egress of Yersiniae from Epithelial Cells. Cell. Microbiol. 2019, 21, e13046. [Google Scholar] [CrossRef]
- David, L.; Taieb, F.; Pénary, M.; Bordignon, P.-J.; Planès, R.; Bagayoko, S.; Duplan-Eche, V.; Meunier, E.; Oswald, E. Outer Membrane Vesicles Produced by Pathogenic Strains of Escherichia coli Block Autophagic Flux and Exacerbate Inflammasome Activation. Autophagy 2022, 18, 2913–2925. [Google Scholar] [CrossRef]
- Harris, J.; Lang, T.; Thomas, J.P.W.; Sukkar, M.B.; Nabar, N.R.; Kehrl, J.H. Autophagy and Inflammasomes. Mol. Immunol. 2017, 86, 10–15. [Google Scholar] [CrossRef]
- Krakauer, T. Inflammasomes, Autophagy, and Cell Death: The Trinity of Innate Host Defense against Intracellular Bacteria. Mediat. Inflamm. 2019, 2019, 2471215. [Google Scholar] [CrossRef]
- Saitoh, T.; Fujita, N.; Jang, M.H.; Uematsu, S.; Yang, B.-G.; Satoh, T.; Omori, H.; Noda, T.; Yamamoto, N.; Komatsu, M.; et al. Loss of the Autophagy Protein Atg16L1 Enhances Endotoxin-Induced IL-1β Production. Nature 2008, 456, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Schroder, K.; Kanneganti, T.-D.; Shao, F.; Broz, P. Mechanisms and Consequences of Inflammasome Activation. J. Mol. Biol. 2018, 430, 131–132. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Tang, B.; Jia, Y.; Zhu, P.; Zhuang, Y.; Fang, Y.; Li, Q.; Wang, K.; Zhang, W.; Guo, G.; et al. Helicobacter pylori CagA Protein Negatively Regulates Autophagy and Promotes Inflammatory Response via C-Met-PI3K/Akt-MTOR Signaling Pathway. Front. Cell. Infect. Microbiol. 2017, 7, 417. [Google Scholar] [CrossRef]
- Chargui, A.; Cesaro, A.; Mimouna, S.; Fareh, M.; Brest, P.; Naquet, P.; Darfeuille-Michaud, A.; Hébuterne, X.; Mograbi, B.; Vouret-Craviari, V.; et al. Subversion of Autophagy in Adherent Invasive Escherichia coli-Infected Neutrophils Induces Inflammation and Cell Death. PLoS ONE 2012, 7, e51727. [Google Scholar] [CrossRef] [PubMed]
- Vigneron, C.; Py, B.F.; Monneret, G.; Venet, F. The Double Sides of NLRP3 Inflammasome Activation in Sepsis. Clin. Sci. 2023, 137, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Eren, E.; Planès, R.; Bagayoko, S.; Bordignon, P.-J.; Chaoui, K.; Hessel, A.; Santoni, K.; Pinilla, M.; Lagrange, B.; Burlet-Schiltz, O.; et al. Irgm2 and Gate-16 Cooperatively Dampen Gram-Negative Bacteria-Induced Caspase-11 Response. EMBO Rep. 2020, 21, e50829. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Lin, S.; Chen, Z.; Qin, B.; He, Z.; Cheng, M.; Sun, M.; Sun, J. Bacteria-Driven Bio-Therapy: From Fundamental Studies to Clinical Trials. Nano Today 2023, 48, 101731. [Google Scholar] [CrossRef]
- Yang, J.; Shin, T.-S.; Kim, J.S.; Jee, Y.-K.; Kim, Y.-K. A New Horizon of Precision Medicine: Combination of the Microbiome and Extracellular Vesicles. Exp. Mol. Med. 2022, 54, 466–482. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pin, C.; David, L.; Oswald, E. Modulation of Autophagy and Cell Death by Bacterial Outer-Membrane Vesicles. Toxins 2023, 15, 502. https://doi.org/10.3390/toxins15080502
Pin C, David L, Oswald E. Modulation of Autophagy and Cell Death by Bacterial Outer-Membrane Vesicles. Toxins. 2023; 15(8):502. https://doi.org/10.3390/toxins15080502
Chicago/Turabian StylePin, Camille, Laure David, and Eric Oswald. 2023. "Modulation of Autophagy and Cell Death by Bacterial Outer-Membrane Vesicles" Toxins 15, no. 8: 502. https://doi.org/10.3390/toxins15080502





