New Insights into Interactions between Mushroom Aegerolysins and Membrane Lipids
Abstract
:1. Introduction
2. Results
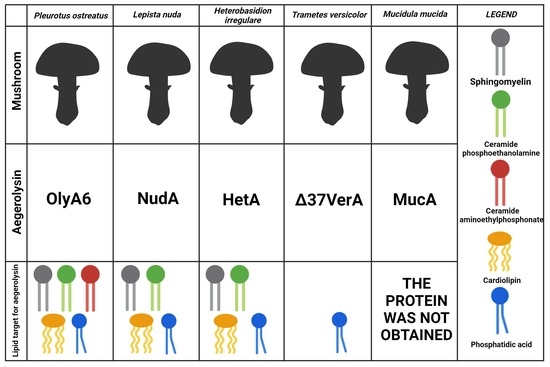
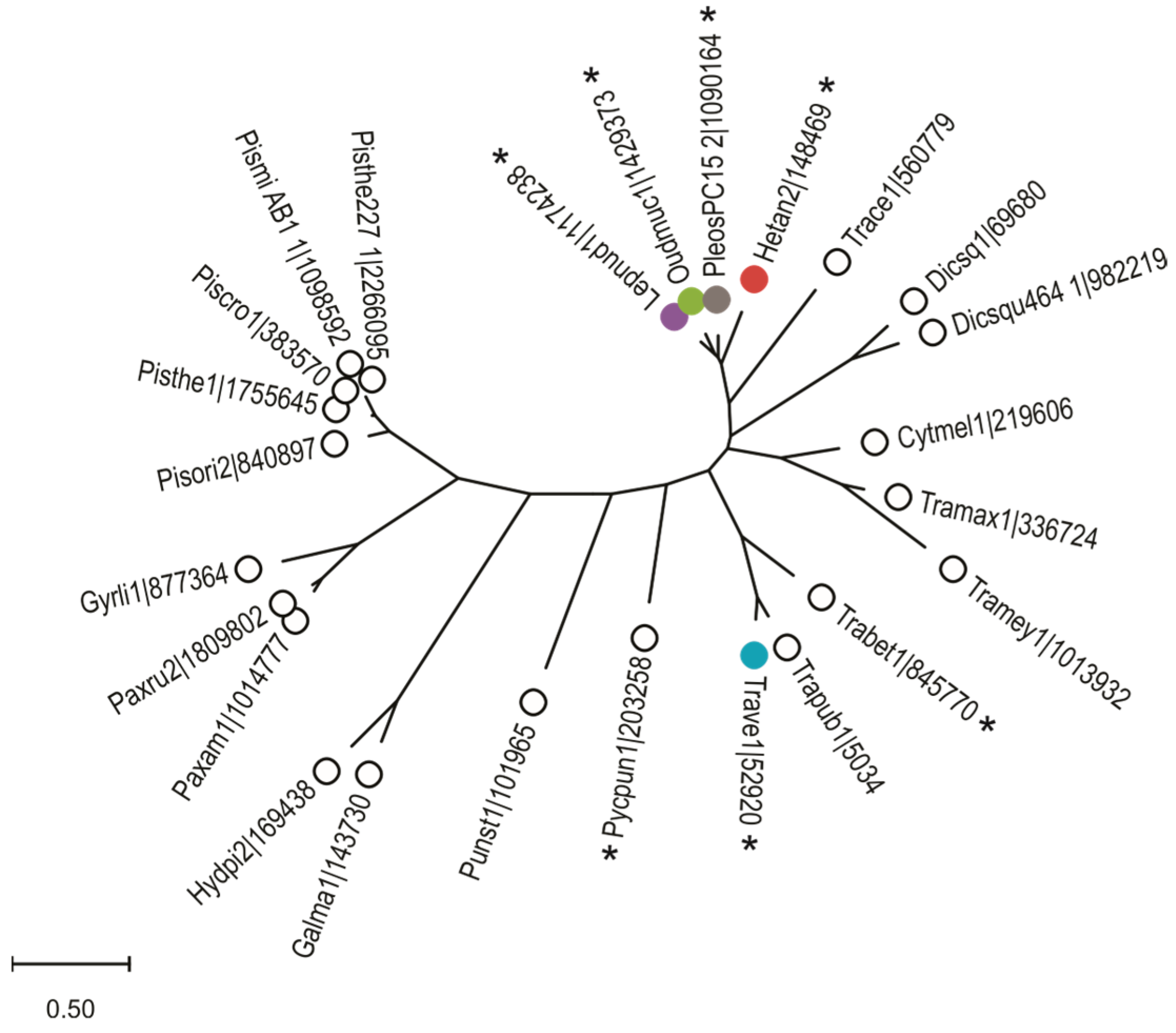
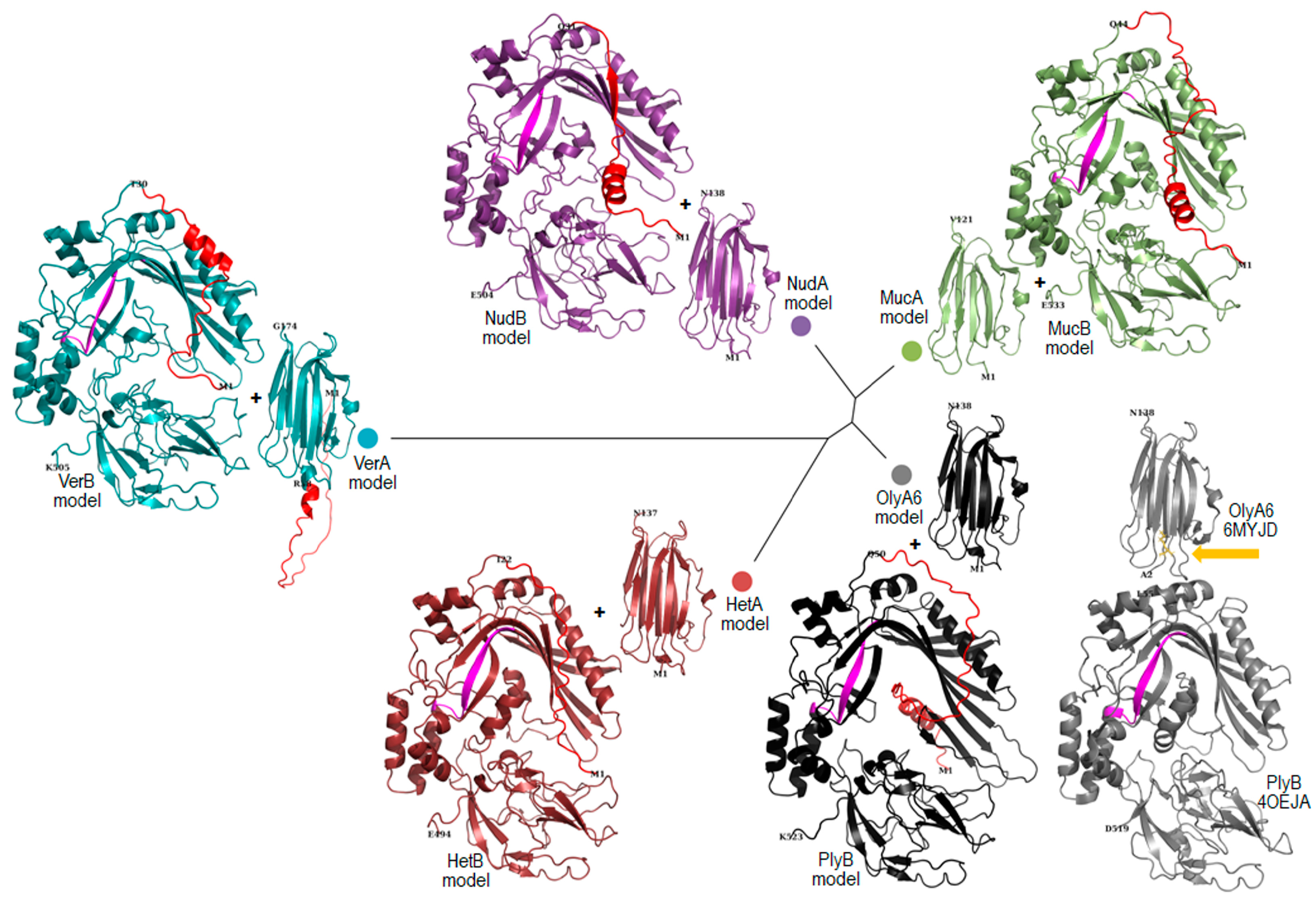
2.1. Bioinformatic Analysis

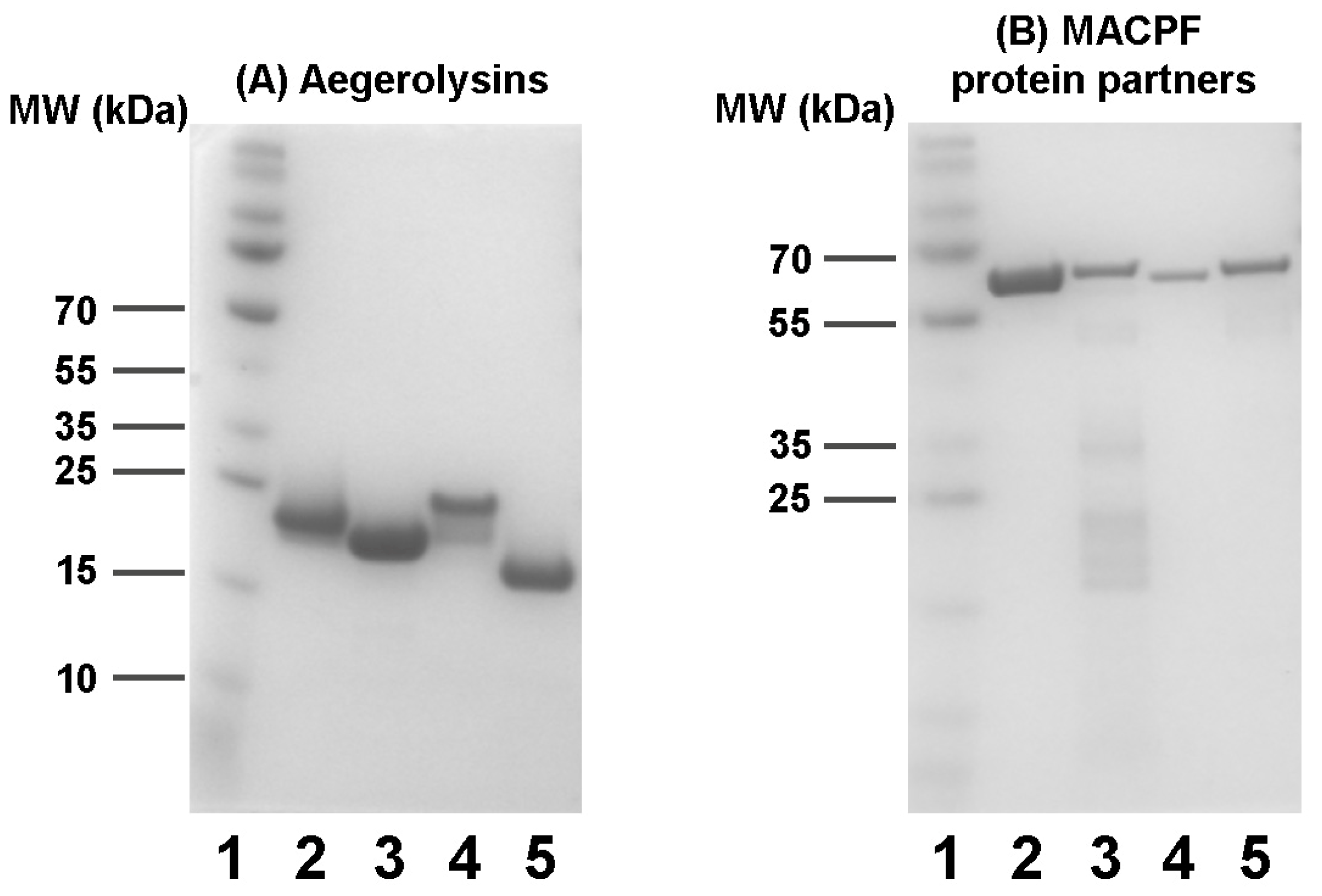
2.2. Production of Recombinant Proteins
2.3. Membrane Binding Studies
2.3.1. Sedimentation Assay
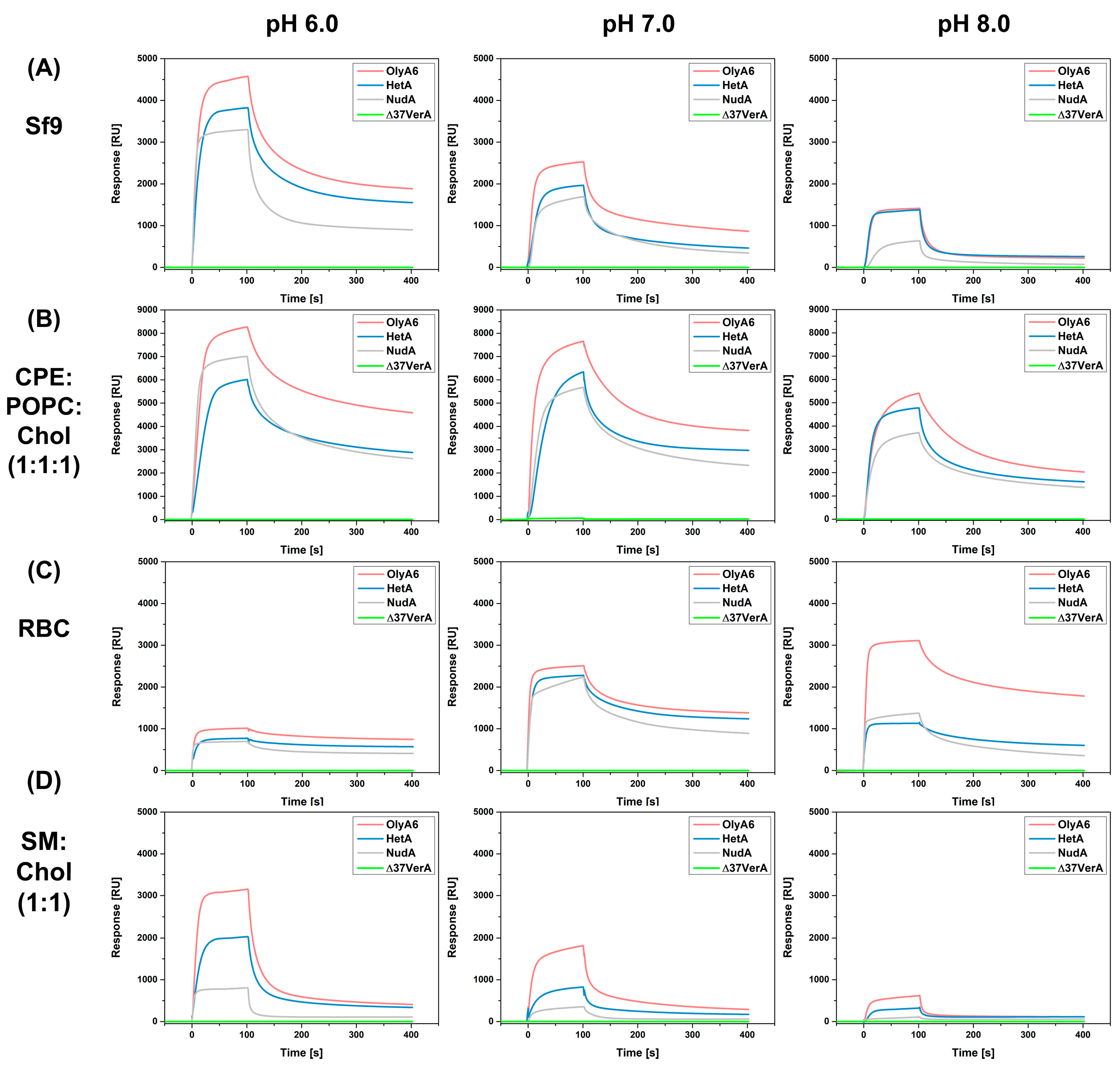
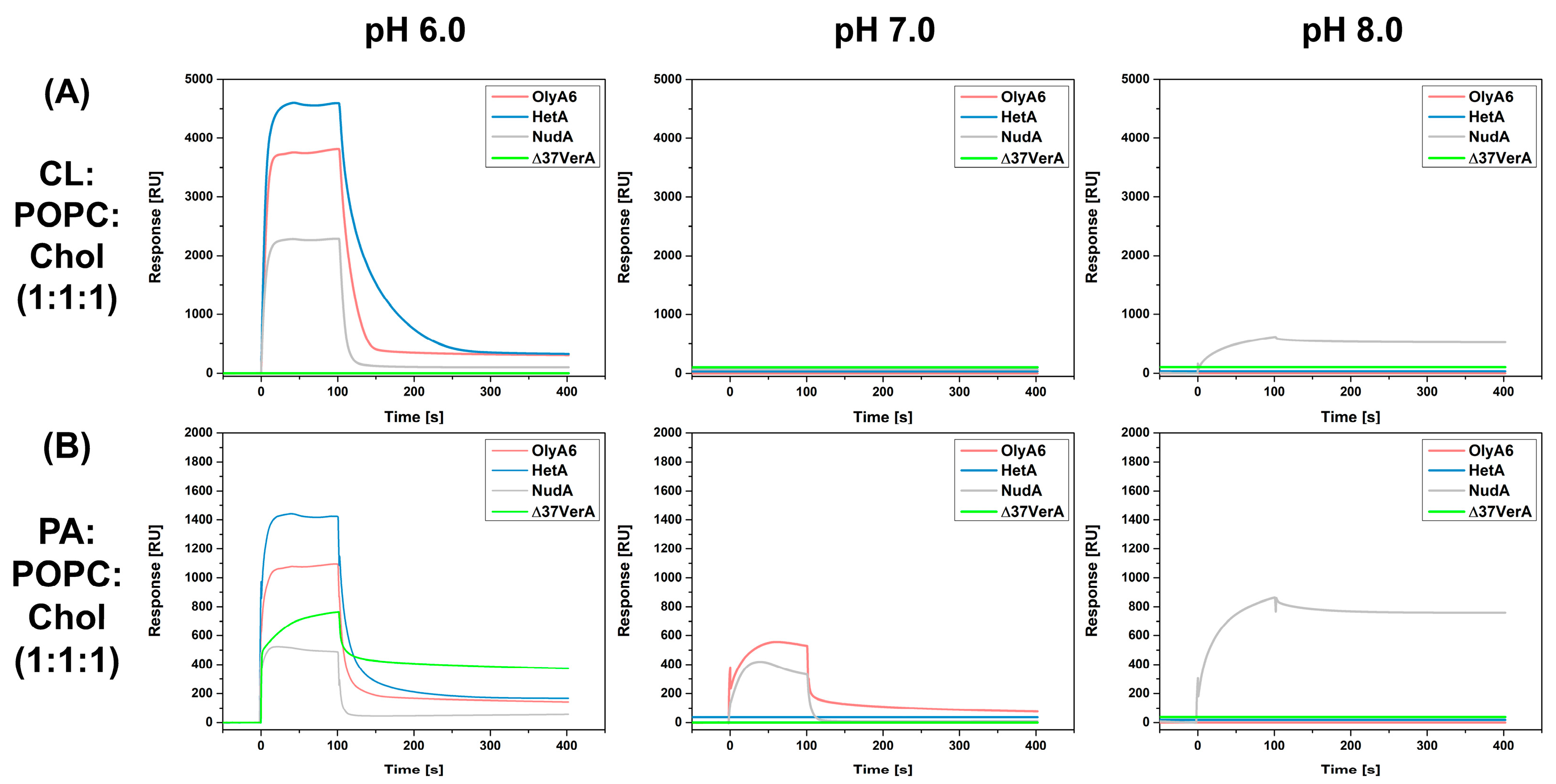
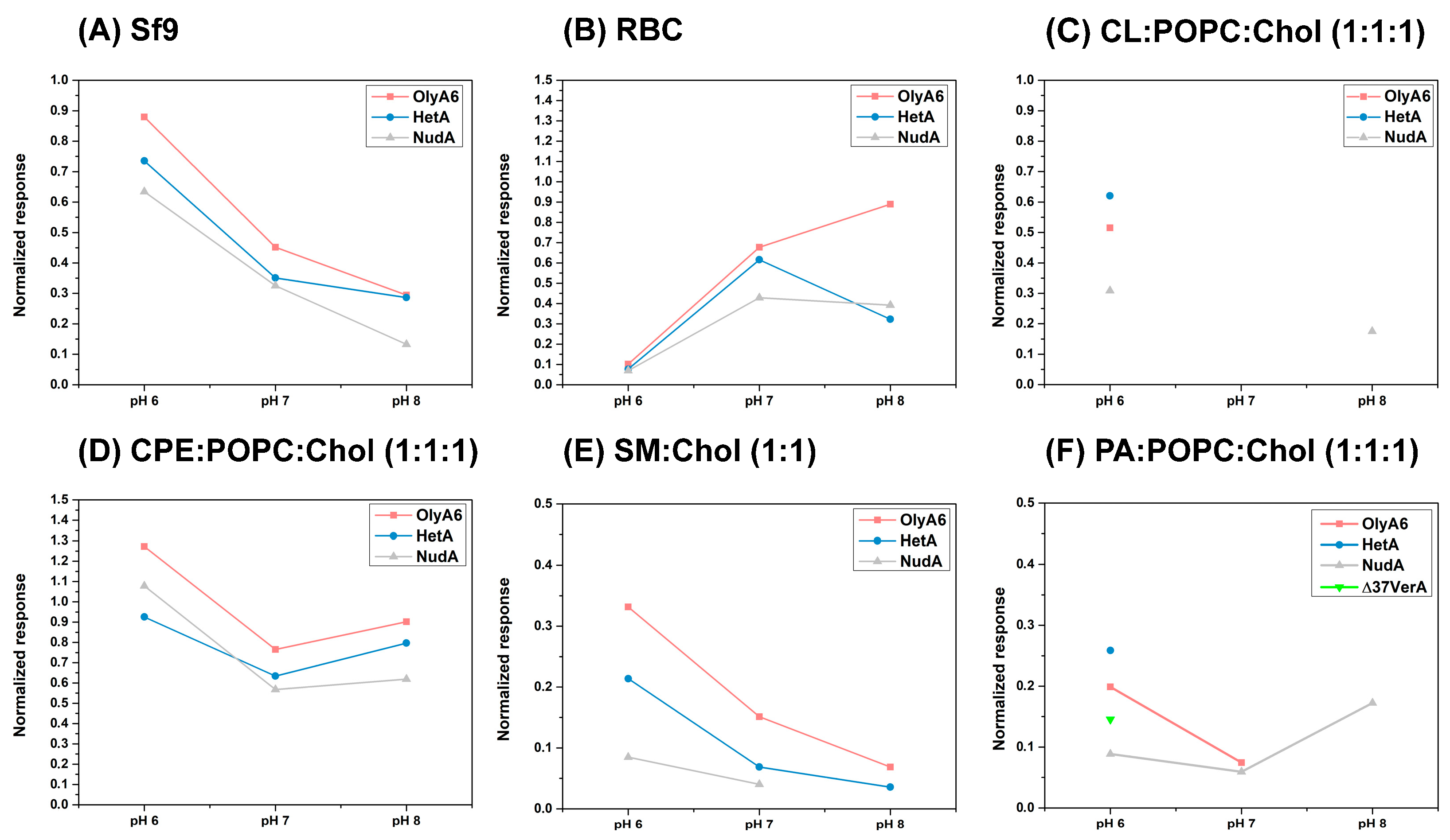
2.3.2. Surface Plasmon Resonance
2.4. Membrane Permeabilization Studies
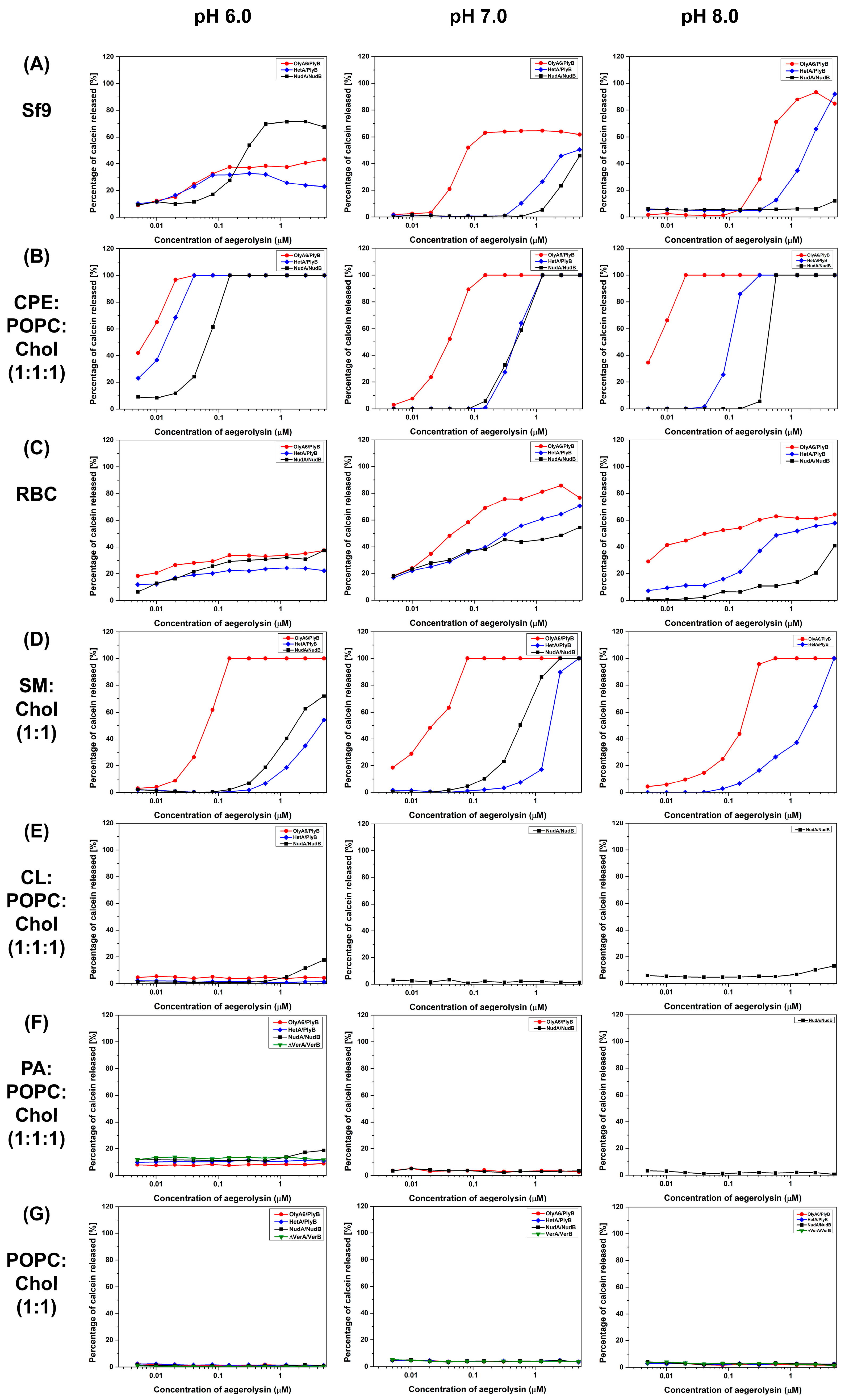
2.4.1. Calcein Release Assay
2.4.2. Hemolytic Activity
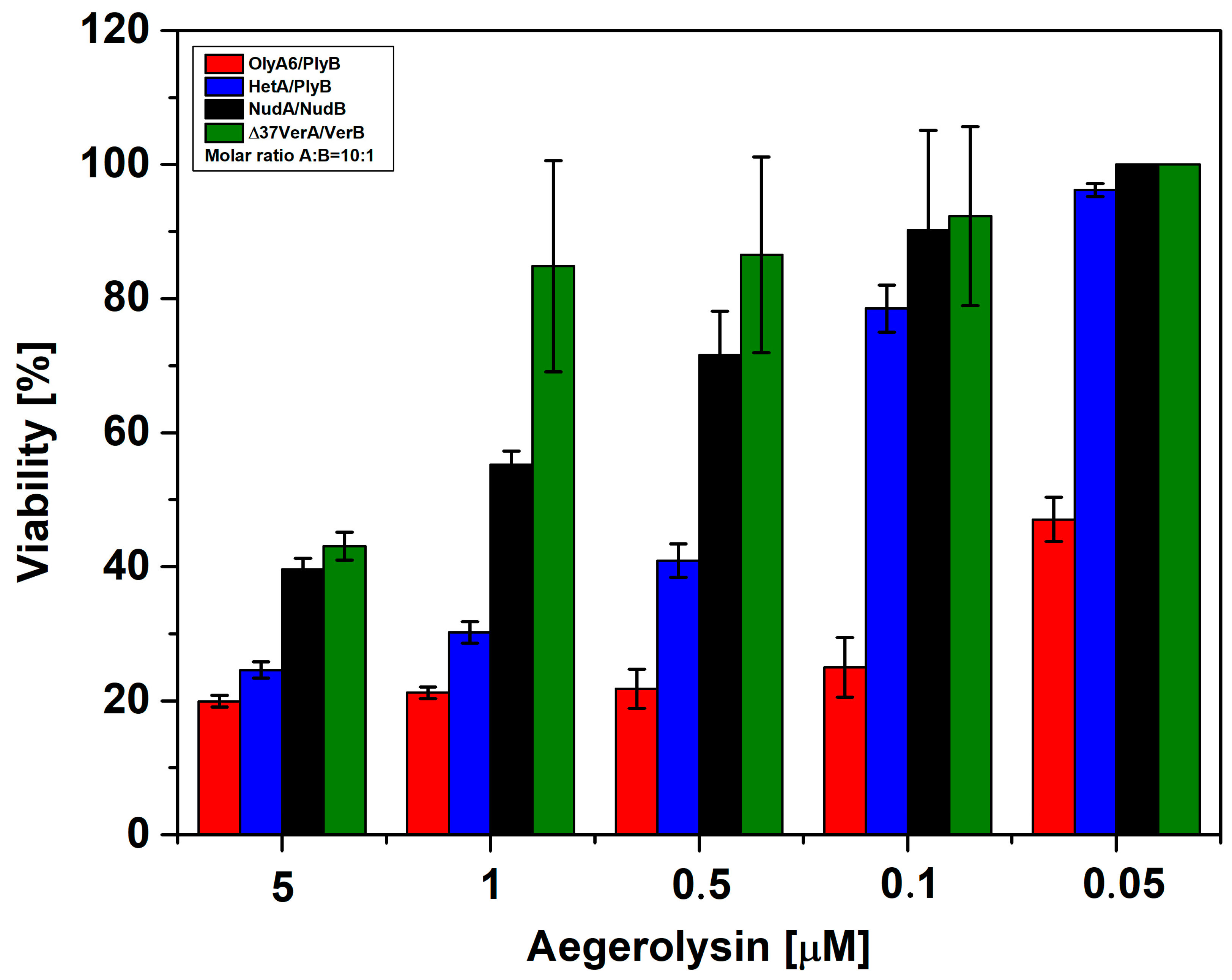
2.4.3. Sf9 Cytotoxicity Studies
3. Discussion
4. Materials and Methods
4.1. Reagents and Materials
4.1.1. Enzymes, Kits, Chemicals, and Cells
4.1.2. Genes, Primers, and Plasmids
4.1.3. Lipids
4.2. Bioinformatics
4.2.1. Identification of Aegerolysin-Coding Basidiomycetes
4.2.2. Phylogenetic Analysis
4.2.3. Calculated Basic Biochemical Characteristic of Aegerolysins and Their MACPF Domain-Containing Protein Partners
4.2.4. Sequence Alignment, Prediction of Protein Secondary Structures, Signal Peptides, and Protein Localisation Prediction
4.2.5. Protein Model Generation
4.2.6. Protein Structure Visualization
4.3. Preparation of Recombinant Proteins
4.4. Preparation of Artificial Lipid Vesicles
4.4.1. Preparation of Lipid Mixtures and Multilamellar Vesicles
4.4.2. Preparation of Large Unilamellar Lipid Vesicles
4.4.3. Preparation of Small Unilamellar Vesicles with Calcein
4.5. Sedimentation Assay
4.6. Surface Plasmon Resonance Measurements
4.7. Calcein Release Assay
4.8. Hemolytic Assay
4.9. Sf9 Cytotoxic Activity
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kraševec, N.; Skočaj, M. Towards Understanding the Function of Aegerolysins. Toxins 2022, 14, 629. [Google Scholar] [CrossRef]
- Tomita, T.; Noguchi, K.; Mimuro, H.; Ukaji, F.; Ito, K.; Sugawara-Tomita, N.; Hashimoto, Y. Pleurotolysin, a Novel Sphingomyelin-Specific Two-Component Cytolysin from the Edible Mushroom Pleurotus Ostreatus, Assembles into a Transmembrane Pore Complex. J. Biol. Chem. 2004, 279, 26975–26982. [Google Scholar] [CrossRef]
- Ota, K.; Leonardi, A.; Mikelj, M.; Skočaj, M.; Wohlschlager, T.; Künzler, M.; Aebi, M.; Narat, M.; Križaj, I.; Anderluh, G.; et al. Membrane Cholesterol and Sphingomyelin, and Ostreolysin A Are Obligatory for Pore-Formation by a MACPF/CDC-like Pore-Forming Protein, Pleurotolysin B. Biochimie 2013, 95, 1855–1864. [Google Scholar] [CrossRef]
- Bhat, H.B.; Ishitsuka, R.; Inaba, T.; Murate, M.; Abe, M.; Makino, A.; Kohyama-Koganeya, A.; Nagao, K.; Kurahashi, A.; Kishimoto, T.; et al. Evaluation of Aegerolysins as Novel Tools to Detect and Visualize Ceramide Phosphoethanolamine, a Major Sphingolipid in Invertebrates. FASEB J. 2015, 29, 3920–3934. [Google Scholar] [CrossRef] [PubMed]
- Sakihara, T.; Takiguchi, N.; Uzawa, H.; Serizawa, R.; Kobayashi, T. Erylysin A Inhibits Cytokinesis in Escherichia Coli by Binding with Cardiolipin. J. Biochem. 2021, 170, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Balbi, T.; Trenti, F.; Panevska, A.; Bajc, G.; Guella, G.; Ciacci, C.; Canonico, B.; Canesi, L.; Sepčić, K. Ceramide Aminoethylphosphonate as a New Molecular Target for Pore-Forming Aegerolysin-Based Protein Complexes. Front. Mol. Biosci. 2022, 9, 902706. [Google Scholar] [CrossRef] [PubMed]
- Yamaji-Hasegawa, A.; Hullin-Matsuda, F.; Greimel, P.; Kobayashi, T. Pore-Forming Toxins: Properties, Diversity, and Uses as Tools to Image Sphingomyelin and Ceramide Phosphoethanolamine. Biochim. Biophys. Acta (BBA)—Biomembr. 2016, 1858, 576–592. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, T.; Ishitsuka, R.; Kobayashi, T. Detectors for Evaluating the Cellular Landscape of Sphingomyelin- and Cholesterol-Rich Membrane Domains. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2016, 1861, 812–829. [Google Scholar] [CrossRef]
- Hullin-Matsuda, F.; Makino, A.; Murate, M.; Kobayashi, T. Probing Phosphoethanolamine-Containing Lipids in Membranes with Duramycin/Cinnamycin and Aegerolysin Proteins. Biochimie 2016, 130, 81–90. [Google Scholar] [CrossRef]
- Hullin-Matsuda, F.; Murate, M.; Kobayashi, T. Protein Probes to Visualize Sphingomyelin and Ceramide Phosphoethanolamine. Chem. Phys. Lipids 2018, 216, 132–141. [Google Scholar] [CrossRef]
- Grundner, M.; Panevska, A.; Sepčić, K.; Skočaj, M. What Can Mushroom Proteins Teach Us about Lipid Rafts? Membranes 2021, 11, 264. [Google Scholar] [CrossRef] [PubMed]
- Skočaj, M.; Resnik, N.; Grundner, M.; Ota, K.; Rojko, N.; Hodnik, V.; Anderluh, G.; Sobota, A.; Maček, P.; Veranič, P.; et al. Tracking Cholesterol/Sphingomyelin-Rich Membrane Domains with the Ostreolysin A-mCherry Protein. PLoS ONE 2014, 9, e92783. [Google Scholar] [CrossRef]
- Shibata, T.; Kudou, M.; Hoshi, Y.; Kudo, A.; Nanashima, N.; Miyairi, K. Isolation and Characterization of a Novel Two-Component Hemolysin, Erylysin A and B, from an Edible Mushroom, Pleurotus Eryngii. Toxicon 2010, 56, 1436–1442. [Google Scholar] [CrossRef]
- Panevska, A.; Hodnik, V.; Skočaj, M.; Novak, M.; Modic, Š.; Pavlic, I.; Podržaj, S.; Zarić, M.; Resnik, N.; Maček, P.; et al. Pore-Forming Protein Complexes from Pleurotus Mushrooms Kill Western Corn Rootworm and Colorado Potato Beetle through Targeting Membrane Ceramide Phosphoethanolamine. Sci. Rep. 2019, 9, 5073. [Google Scholar] [CrossRef]
- Milijaš Jotić, M.; Panevska, A.; Iacovache, I.; Kostanjšek, R.; Mravinec, M.; Skočaj, M.; Zuber, B.; Pavšič, A.; Razinger, J.; Modic, Š.; et al. Dissecting out the Molecular Mechanism of Insecticidal Activity of Ostreolysin A6/Pleurotolysin B Complexes on Western Corn Rootworm. Toxins 2021, 13, 455. [Google Scholar] [CrossRef]
- Resnik, N.; Repnik, U.; Kreft, M.E.; Sepčić, K.; Maček, P.; Turk, B.; Veranič, P. Highly Selective Anti-Cancer Activity of Cholesterol-Interacting Agents Methyl-β-Cyclodextrin and Ostreolysin A/Pleurotolysin B Protein Complex on Urothelial Cancer Cells. PLoS ONE 2015, 10, e0137878. [Google Scholar] [CrossRef]
- De, J.; Nandi, S.; Acharya, K. A Review on Blewit Mushrooms (Lepista sp.) Transition from Farm to Pharm. J. Food Process. Preserv. 2022, 46, e17028. [Google Scholar] [CrossRef]
- Niego, A.G.; Raspé, O.; Thongklang, N.; Charoensup, R.; Lumyong, S.; Stadler, M.; Hyde, K.D. Taxonomy, Diversity and Cultivation of the Oudemansielloid/Xeruloid Taxa Hymenopellis, Mucidula, Oudemansiella, and Xerula with Respect to Their Bioactivities: A Review. J. Fungi 2021, 7, 51. [Google Scholar] [CrossRef]
- Chen, J.-J.; Cui, B.-K.; Zhou, L.-W.; Korhonen, K.; Dai, Y.-C. Phylogeny, Divergence Time Estimation, and Biogeography of the Genus Heterobasidion (Basidiomycota, Russulales). Fungal Divers. 2015, 71, 185–200. [Google Scholar] [CrossRef]
- Coyle, D.R.; Klepzig, K.D.; Koch, F.H.; Morris, L.A.; Nowak, J.T.; Oak, S.W.; Otrosina, W.J.; Smith, W.D.; Gandhi, K.J.K. A Review of Southern Pine Decline in North America. For. Ecol. Manag. 2015, 349, 134–148. [Google Scholar] [CrossRef]
- Pointing, S. Feasibility of Bioremediation by White-Rot Fungi. Appl. Microbiol. Biotechnol. 2001, 57, 20–33. [Google Scholar] [CrossRef]
- Habtemariam, S. Trametes Versicolor (Synn. Coriolus versicolor) Polysaccharides in Cancer Therapy: Targets and Efficacy. Biomedicines 2020, 8, 135. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm Portal: Gearing up for 1000 Fungal Genomes. Nucleic Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef] [PubMed]
- MycoCosm—The Fungal Genomics Resouce (DOE Joint Genome Institute). Available online: https://mycocosm.jgi.doe.gov/mycocosm/home (accessed on 27 July 2023).
- NCBI Taxonomy. Available online: https://www.ncbi.nlm.nih.gov/taxonomy (accessed on 27 July 2023).
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The Rapid Generation of Mutation Data Matrices from Protein Sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Kraševec, N.; Novak, M.; Barat, S.; Skočaj, M.; Sepčić, K.; Anderluh, G. Unconventional Secretion of Nigerolysins A from Aspergillus Species. Microorganisms 2020, 8, 1973. [Google Scholar] [CrossRef] [PubMed]
- Anderluh, G.; Kisovec, M.; Kraševec, N.; Gilbert, R.J.C. Distribution of MACPF/CDC Proteins. Sub-Cell. Biochem. 2014, 80, 7–30. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- The Molecular Graphics System PyMOL (Schrödinger, LLC). Available online: https://pymol.org/2/ (accessed on 4 October 2021).
- Endapally, S.; Frias, D.; Grzemska, M.; Gay, A.; Tomchick, D.R.; Radhakrishnan, A. Molecular Discrimination between Two Conformations of Sphingomyelin in Plasma Membranes. Cell 2019, 176, 1040–1053.e17. [Google Scholar] [CrossRef]
- Lukoyanova, N.; Kondos, S.C.; Farabella, I.; Law, R.H.P.; Reboul, C.F.; Caradoc-Davies, T.T.; Spicer, B.A.; Kleifeld, O.; Traore, D.A.K.; Ekkel, S.M.; et al. Conformational Changes during Pore Formation by the Perforin-Related Protein Pleurotolysin. PLOS Biol. 2015, 13, e1002049. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Eisenberg, D.; Schwarz, E.; Komaromy, M.; Wall, R. Analysis of Membrane and Surface Protein Sequences with the Hydrophobic Moment Plot. J. Mol. Biol. 1984, 179, 125–142. [Google Scholar] [CrossRef]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS Biomolecular Solvation Software Suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 Predicts All Five Types of Signal Peptides Using Protein Language Models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Thumuluri, V.; Almagro Armenteros, J.J.; Johansen, A.R.; Nielsen, H.; Winther, O. DeepLoc 2.0: Multi-Label Subcellular Localization Prediction Using Protein Language Models. Nucleic Acids Res. 2022, 50, W228–W234. [Google Scholar] [CrossRef]
- Novak, M.; Krpan, T.; Panevska, A.; Shewell, L.K.; Day, C.J.; Jennings, M.P.; Guella, G.; Sepčić, K. Binding Specificity of Ostreolysin A6 towards Sf9 Insect Cell Lipids. Biochim. Biophys. Acta (BBA)—Biomembr. 2020, 1862, 183307. [Google Scholar] [CrossRef]
- Bernheimer, A.W.; Avigad, L.S. A Cytolytic Protein from the Edible Mushroom, Pleurotus Ostreatus. Biochim. Biophys. Acta 1979, 585, 451–461. [Google Scholar] [CrossRef]
- Panevska, A.; Razinger, J.; Sepčić, K.; Maček, P.; Skočaj, M.; Modic, S.; Novak, M.; Butala, M.; Hodnik, V.; Grundner, M.; et al. New Bio-Pesticides for Controlling Plant Pests. U.S. Patent Application No. 16/649,385, 6 August 2020. [Google Scholar]
- Ivanov, I.T. Low pH-Induced Hemolysis of Erythrocytes Is Related to the Entry of the Acid into Cytosole and Oxidative Stress on Cellular Membranes. Biochim. Biophys. Acta (BBA)—Biomembr. 1999, 1415, 349–360. [Google Scholar] [CrossRef]
- Olofsson, G.; Sparr, E. Ionization Constants pKa of Cardiolipin. PLoS ONE 2013, 8, e73040. [Google Scholar] [CrossRef] [PubMed]
- Sathappa, M.; Alder, N.N. The Ionization Properties of Cardiolipin and Its Variants in Model Bilayers. Biochim. Biophys. Acta (BBA)—Biomembr. 2016, 1858, 1362–1372. [Google Scholar] [CrossRef] [PubMed]
- Thakur, R.; Naik, A.; Panda, A.; Raghu, P. Regulation of Membrane Turnover by Phosphatidic Acid: Cellular Functions and Disease Implications. Front. Cell Dev. Biol. 2019, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.L.; Cestra, G.; Shui, G.; Kuhrs, A.; Schittenhelm, R.B.; Hafen, E.; van der Goot, F.G.; Robinett, C.C.; Gatti, M.; Gonzalez-Gaitan, M.; et al. Biochemical Membrane Lipidomics during Drosophila Development. Dev. Cell 2013, 24, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, R.; Nagao, K.; Taniuchi, K.; Tsuchiya, M.; Kato, U.; Hara, Y.; Inaba, T.; Kobayashi, T.; Sasaki, Y.; Akiyoshi, K.; et al. Development of a Novel Tetravalent Synthetic Peptide That Binds to Phosphatidic Acid. PLoS ONE 2015, 10, e0131668. [Google Scholar] [CrossRef] [PubMed]
- Berne, S.; Sepcić, K.; Anderluh, G.; Turk, T.; Macek, P.; Poklar Ulrih, N. Effect of pH on the pore forming activity and conformational stability of ostreolysin, a lipid raft-binding protein from the edible mushroom Pleurotus ostreatus. Biochemistry 2005, 44, 11137–11147. [Google Scholar] [CrossRef] [PubMed]
- Kaiser-Alexnat, R. Protease Activities in the Midgut of Western Corn Rootworm (Diabrotica Virgifera Virgifera LeConte). J. Invertebr. Pathol. 2009, 100, 169–174. [Google Scholar] [CrossRef]
- Panevska, A.; Glavan, G.; Jemec Kokalj, A.; Kukuljan, V.; Trobec, T.; Žužek, M.C.; Vrecl, M.; Drobne, D.; Frangež, R.; Sepčić, K. Effects of Bioinsecticidal Aegerolysin-Based Cytolytic Complexes on Non-Target Organisms. Toxins 2021, 13, 457. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A Protein Secondary Structure Prediction Server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Sepčić, K.; Berne, S.; Potrich, C.; Turk, T.; Maček, P.; Menestrina, G. Interaction of Ostreolysin, a Cytolytic Protein from the Edible Mushroom Pleurotus Ostreatus, with Lipid Membranes and Modulation by Lysophospholipids. Eur. J. Biochem. 2003, 270, 1199–1210. [Google Scholar] [CrossRef]
- Belmonte, G.; Pederzolli, C.; Maček, P.; Menestrina, G. Pore Formation by the Sea Anemone Cytolysin Equinatoxin II in Red Blood Cells and Model Lipid Membranes. J. Membr. Biol. 1993, 131, 11–22. [Google Scholar] [CrossRef]
- Riley, R.; Salamov, A.A.; Brown, D.W.; Nagy, L.G.; Floudas, D.; Held, B.W.; Levasseur, A.; Lombard, V.; Morin, E.; Otillar, R.; et al. Extensive Sampling of Basidiomycete Genomes Demonstrates Inadequacy of the White-Rot/Brown-Rot Paradigm for Wood Decay Fungi. Proc. Natl. Acad. Sci. USA 2014, 111, 9923–9928. [Google Scholar] [CrossRef]
- Ruiz-Dueñas, F.J.; Barrasa, J.M.; Sánchez-García, M.; Camarero, S.; Miyauchi, S.; Serrano, A.; Linde, D.; Babiker, R.; Drula, E.; Ayuso-Fernández, I.; et al. Genomic Analysis Enlightens Agaricales Lifestyle Evolution and Increasing Peroxidase Diversity. Mol. Biol. Evol. 2021, 38, 1428–1446. [Google Scholar] [CrossRef]
- Alfaro, M.; Castanera, R.; Lavín, J.L.; Grigoriev, I.V.; Oguiza, J.A.; Ramírez, L.; Pisabarro, A.G. Comparative and Transcriptional Analysis of the Predicted Secretome in the Lignocellulose-Degrading Basidiomycete Fungus Pleurotus ostreatus. Environ. Microbiol. 2016, 18, 4710–4726. [Google Scholar] [CrossRef] [PubMed]
- Varga, T.; Krizsán, K.; Földi, C.; Dima, B.; Sánchez-García, M.; Sánchez-Ramírez, S.; Szöllősi, G.J.; Szarkándi, J.G.; Papp, V.; Albert, L.; et al. Megaphylogeny Resolves Global Patterns of Mushroom Evolution. Nat. Ecol. Evol. 2019, 3, 668–678. [Google Scholar] [CrossRef]
- Miyauchi, S.; Kiss, E.; Kuo, A.; Drula, E.; Kohler, A.; Sánchez-García, M.; Morin, E.; Andreopoulos, B.; Barry, K.W.; Bonito, G.; et al. Large-Scale Genome Sequencing of Mycorrhizal Fungi Provides Insights into the Early Evolution of Symbiotic Traits. Nat. Commun. 2020, 11, 5125. [Google Scholar] [CrossRef] [PubMed]
- Kohler, A.; Kuo, A.; Nagy, L.G.; Morin, E.; Barry, K.W.; Buscot, F.; Canbäck, B.; Choi, C.; Cichocki, N.; Clum, A.; et al. Convergent Losses of Decay Mechanisms and Rapid Turnover of Symbiosis Genes in Mycorrhizal Mutualists. Nat. Genet. 2015, 47, 410–415. [Google Scholar] [CrossRef]
- Plett, J.M.; Miyauchi, S.; Morin, E.; Plett, K.; Wong-Bajracharya, J.; de Freitas Pereira, M.; Kuo, A.; Henrissat, B.; Drula, E.; Wojtalewicz, D.; et al. Speciation Underpinned by Unexpected Molecular Diversity in the Mycorrhizal Fungal Genus Pisolithus. Mol. Biol. Evol. 2023, 40, msad045. [Google Scholar] [CrossRef] [PubMed]
- Balasundaram, S.V.; Hess, J.; Durling, M.B.; Moody, S.C.; Thorbek, L.; Progida, C.; LaButti, K.; Aerts, A.; Barry, K.; Grigoriev, I.V.; et al. The Fungus That Came in from the Cold: Dry Rot’s Pre-Adapted Ability to Invade Buildings. ISME J. 2018, 12, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Floudas, D.; Binder, M.; Riley, R.; Barry, K.; Blanchette, R.A.; Henrissat, B.; Martinez, A.T.; Otillar, R.; Spatafora, J.W.; Yadav, J.S.; et al. The Paleozoic Origin of Enzymatic Lignin Decomposition Reconstructed from 31 Fungal Genomes. Science 2012, 336, 1715–1719. [Google Scholar] [CrossRef]
- Hage, H.; Miyauchi, S.; Virágh, M.; Drula, E.; Min, B.; Chaduli, D.; Navarro, D.; Favel, A.; Norest, M.; Lesage-Meessen, L.; et al. Gene Family Expansions and Transcriptome Signatures Uncover Fungal Adaptations to Wood Decay. Environ. Microbiol. 2021, 23, 5716–5732. [Google Scholar] [CrossRef] [PubMed]
- Casado López, S.; Peng, M.; Daly, P.; Andreopoulos, B.; Pangilinan, J.; Lipzen, A.; Riley, R.; Ahrendt, S.; Ng, V.; Barry, K.; et al. Draft Genome Sequences of Three Monokaryotic Isolates of the White-Rot Basidiomycete Fungus Dichomitus Squalens. Microbiol. Resour. Announc. 2019, 8, e00264-19. [Google Scholar] [CrossRef]
- Granchi, Z.; Peng, M.; Chi-A-Woeng, T.; de Vries, R.P.; Hildén, K.; Mäkelä, M.R. Genome Sequence of the Basidiomycete White-Rot Fungus Trametes Pubescens FBCC735. Genome Announc. 2017, 5, e01643-16. [Google Scholar] [CrossRef] [PubMed]
- Olson, Å.; Aerts, A.; Asiegbu, F.; Belbahri, L.; Bouzid, O.; Broberg, A.; Canbäck, B.; Coutinho, P.M.; Cullen, D.; Dalman, K.; et al. Insight into Trade-off between Wood Decay and Parasitism from the Genome of a Fungal Forest Pathogen. New Phytol. 2012, 194, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- Meinhardt, L.W.; Costa, G.G.; Thomazella, D.P.; Teixeira, P.J.P.; Carazzolle, M.; Schuster, S.C.; Carlson, J.E.; Guiltinan, M.J.; Mieczkowski, P.; Farmer, A.; et al. Genome and Secretome Analysis of the Hemibiotrophic Fungal Pathogen, Moniliophthora Roreri, Which Causes Frosty Pod Rot Disease of Cacao: Mechanisms of the Biotrophic and Necrotrophic Phases. BMC Genom. 2014, 15, 164. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.D.; Perkins, A.D.; Sonstegard, T.S.; Schroeder, S.G.; Burgess, S.C.; Diehl, S.V. Short-Read Sequencing for Genomic Analysis of the Brown Rot Fungus Fibroporia Radiculosa. Appl. Environ. Microbiol. 2012, 78, 2272–2281. [Google Scholar] [CrossRef] [PubMed]
- Steindorff, A.S.; Carver, A.; Calhoun, S.; Stillman, K.; Liu, H.; Lipzen, A.; He, G.; Yan, M.; Pangilinan, J.; LaButti, K.; et al. Comparative Genomics of Pyrophilous Fungi Reveals a Link between Fire Events and Developmental Genes. Environ. Microbiol. 2021, 23, 99–109. [Google Scholar] [CrossRef]
- Bao, D.; Gong, M.; Zheng, H.; Chen, M.; Zhang, L.; Wang, H.; Jiang, J.; Wu, L.; Zhu, Y.; Zhu, G.; et al. Sequencing and Comparative Analysis of the Straw Mushroom (Volvariella volvacea) Genome. PLoS ONE 2013, 8, e58294. [Google Scholar] [CrossRef]
- Wu, B.; Xu, Z.; Knudson, A.; Carlson, A.; Chen, N.; Kovaka, S.; LaButti, K.; Lipzen, A.; Pennachio, C.; Riley, R.; et al. Genomics and Development of Lentinus Tigrinus: A White-Rot Wood-Decaying Mushroom with Dimorphic Fruiting Bodies. Genome Biol. Evol. 2018, 10, 3250–3261. [Google Scholar] [CrossRef]
- Looney, B.; Miyauchi, S.; Morin, E.; Drula, E.; Courty, P.E.; Kohler, A.; Kuo, A.; LaButti, K.; Pangilinan, J.; Lipzen, A.; et al. Evolutionary Transition to the Ectomycorrhizal Habit in the Genomes of a Hyperdiverse Lineage of Mushroom-Forming Fungi. New Phytol. 2022, 233, 2294–2309. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Miyauchi, S.; Morin, E.; Kuo, A.; Drula, E.; Varga, T.; Kohler, A.; Feng, B.; Cao, Y.; Lipzen, A.; et al. Evolutionary Innovations through Gain and Loss of Genes in the Ectomycorrhizal Boletales. New Phytol. 2022, 233, 1383–1400. [Google Scholar] [CrossRef]
- Miettinen, O.; Riley, R.; Barry, K.; Cullen, D.; de Vries, R.P.; Hainaut, M.; Hatakka, A.; Henrissat, B.; Hildén, K.; Kuo, R.; et al. Draft Genome Sequence of the White-Rot Fungus Obba Rivulosa 3A-2. Genome Announc. 2016, 4, e00976-16. [Google Scholar] [CrossRef] [PubMed]
- Mondego, J.M.C.; Carazzolle, M.F.; Costa, G.G.L.; Formighieri, E.F.; Parizzi, L.P.; Rincones, J.; Cotomacci, C.; Carraro, D.M.; Cunha, A.F.; Carrer, H.; et al. A Genome Survey of Moniliophthora Perniciosa Gives New Insights into Witches’ Broom Disease of Cacao. BMC Genom. 2008, 9, 548. [Google Scholar] [CrossRef] [PubMed]
- Ahrendt, S.R.; Mondo, S.J.; Haridas, S.; Grigoriev, I.V. MycoCosm, the JGI’s Fungal Genome Portal for Comparative Genomic and Multiomics Data Analyses. In Microbial Environmental Genomics (MEG); Martin, F., Uroz, S., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2023; pp. 271–291. ISBN 978-1-07-162871-3. [Google Scholar]
| Protein Mixture (10:1) | Concertation of an Aegerolysin in the Protein Mixture | pH 6.0 | pH 7.0 | pH 8.0 |
|---|---|---|---|---|
| Time of Hemolysis (min) | Time of Hemolysis (min) | Time of Hemolysis (min) | ||
| OlyA6/PlyB | 1 µM of OlyA6 | 0.7 ± 0.0 | 0.7 ± 0.0 | 0.7 ± 0.0 |
| 0.1 µM of OlyA6 | 0.7 ± 0.0 | 4 ± 1.1 | 2 ± 0.0 | |
| 0.01 µM of OlyA6 | 2.7 ± 0.5 | 20.6 ± 2.1 | 26.4 ± 1.4 | |
| NudA/NudB | 1 µM of NudA | 0.7 ± 0.0 | 0.7 ± 0.0 | 2 ± 0.0 |
| 0.1 µM of NudA | 1.3 ± 0.4 | 11.1 ± 1.1 | / | |
| 0.01 µM of NudA | 48.8 ± 3.9 | / | / | |
| Δ37VerA/VerB | 1 µM of Δ37VerA | / | / | / |
| 0.1 µM of Δ37VerA | / | / | / | |
| 0.01 µM of Δ37VerA | / | / | / | |
| HetA/PlyB | 1 µM of HetA | 0.7 ± 0.0 | 1.3 ± 0.3 | 0.7 ± 0.0 |
| 0.1 µM of HetA | 0.7 ± 0.0 | 8.2 ± 1.0 | 5.3 ± 0.9 | |
| 0.01 µM of HetA | 9.8 ± 1.2 | / | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popošek, L.L.; Kraševec, N.; Bajc, G.; Glavač, U.; Hrovatin, M.; Perko, Ž.; Slavič, A.; Pavšič, M.; Sepčić, K.; Skočaj, M. New Insights into Interactions between Mushroom Aegerolysins and Membrane Lipids. Toxins 2024, 16, 143. https://doi.org/10.3390/toxins16030143
Popošek LL, Kraševec N, Bajc G, Glavač U, Hrovatin M, Perko Ž, Slavič A, Pavšič M, Sepčić K, Skočaj M. New Insights into Interactions between Mushroom Aegerolysins and Membrane Lipids. Toxins. 2024; 16(3):143. https://doi.org/10.3390/toxins16030143
Chicago/Turabian StylePopošek, Larisa Lara, Nada Kraševec, Gregor Bajc, Urška Glavač, Matija Hrovatin, Žan Perko, Ana Slavič, Miha Pavšič, Kristina Sepčić, and Matej Skočaj. 2024. "New Insights into Interactions between Mushroom Aegerolysins and Membrane Lipids" Toxins 16, no. 3: 143. https://doi.org/10.3390/toxins16030143