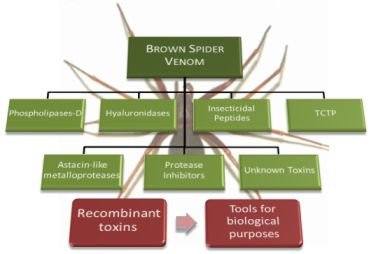
Brown Spider (Loxosceles genus) Venom Toxins: Tools for Biological Purposes
Abstract
:1. The Spiders of Genus Loxosceles and Loxoscelism

2. The Loxosceles Venoms
| Toxins | MW (kDa) | Characteristics and actions described | No. Seq * |
|---|---|---|---|
| Phospholipases-D (SicTox family members, such as LiRecDTs) | 30–35 | Several isoforms with variant features such as:
| 335 |
| Insecticidal peptides | 5–8 | 8 | |
| Metalloproteases | 28–35 | 4 | |
| Hyaluronidases | 41–43 |
| - |
| Serine-proteases | 85–95 | - | |
| Serine/Cysteine protease inhibitors | N.D. | - | |
| TCTP (translationally controlled tumour protein) | ~46 |
| - |
| Lectin-like | N.D. | - Putative features: carbohydrate-binding molecules; involved in extracellular matrix organization, endocytosis, complement activation, etc. [51] | - |
| Alkaline-phosphatase | N.D. | - Degrades the synthetic substrate p-nitrophenyl phosphate[10] | - |
| ATPase | N.D | - ATP hydrolysis [10] | - |
3. The Rational Use of Venom Toxins as Biotechnological Tools
4. Phospholipase-D
5. Hyaluronidase
6. Translationally Controlled Tumor Protein (TCTP)
7. Astacin-Like Metalloproteases
8. Insecticidal Peptides
9. Serine Protease Inhibitors
10. Conclusion
Acknowledgements
References
- Binford, G.J.; Bodner, M.R.; Cordes, M.H.; Baldwin, K.L.; Rynerson, M.R.; Burns, S.N.; Zobel-Thropp, P.A. Molecular evolution, functional variation, and proposed nomenclature of the gene family that includes sphingomyelinase D in sicariid spider venoms. Mol. Biol. Evol. 2009, 26, 547–566. [Google Scholar] [PubMed]
- Platnick, N.I. The World Spider Catalog; American Museum of Natural History: New York, NY, USA, 2008; Version. 9.0. [Google Scholar]
- Binford, G.J.; Callahan, M.S.; Bodner, M.R.; Rynerson, M.R.; Nunez, P.B.; Ellison, C.E.; Duncan, R.P. Phylogenetic relationships of Loxosceles and Sicarius spiders are consistent with Western Gondwanan vicariance. Mol. Phylogenet. Evol. 2008, 49, 538–553. [Google Scholar]
- Appel, M.H.; Bertoni da Silveira, R.; Gremski, W.; Veiga, S.S. Insights into brown spider and loxoscelism. Invertebr. Surviv. J. 2005, 2, 152–158. [Google Scholar]
- Futrell, J.M. Loxoscelism. Am. J. Med. Sci. 1992, 304, 261–267. [Google Scholar]
- da Silva, P.H.; da Silveira, R.B.; Appel, M.H.; Mangili, O.C.; Gremski, W.; Veiga, S.S. Brown spiders and loxoscelism. Toxicon 2004, 44, 693–709. [Google Scholar]
- Hogan, C.J.; Barbaro, K.C.; Winkel, K. Loxoscelism: old obstacles, new directions. Ann. Emerg. Med. 2004, 44, 608–624. [Google Scholar]
- Swanson, D.L.; Vetter, R.S. Loxoscelism. Clinics. Dermatol. 2006, 24, 213–221. [Google Scholar]
- Lung, J.M.; Mallory, S.B. A child with spider bite and glomerulonephritis: A diagnostic challenge. Int. J. Dermatol. 2000, 39, 287–289. [Google Scholar]
- Sales, P.B.; Santoro, M.L. Nucleotidase and DNase activities in Brazilian snake venoms. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2008, 147, 85–95. [Google Scholar]
- Schroeder, F.C.; Taggi, A.E.; Gronquist, M.; Malik, R.U.; Grant, J.B.; Eisner, T.; Meinwald, J. NMR spectroscopic screening of spider venom reveals sulfated nucleosides as major components for the brown recluse and related species. Proc. Natl. Acad. Sci. USA 2008, 105, 14283–14287. [Google Scholar]
- Young, A.R.; Pincus, S.J. Comparison of enzymatic activity from three species of necrotising arachnids in Australia: Loxosceles rufescens, Badumna insignis and Lampona cylindrata. Toxicon 2001, 39, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Barbaro, K.C.; Knysak, I.; Martins, R.; Hogan, C.; Winkel, K. Enzymatic characterization, antigenic cross-reactivity and neutralization of dermonecrotic activity of five Loxosceles spider venoms of medical importance in the Americas. Toxicon 2005, 45, 489–499. [Google Scholar]
- da Silveira, R.B.; Chaim, O.M.; Mangili, O.C.; Gremski, W.; Dietrich, C.P.; Nader, H.B.; Veiga, S.S. Hyaluronidases in Loxosceles intermedia (Brown spider) venom are endo-beta-N-acetyl-d-hexosaminidases hydrolases. Toxicon 2007, 49, 758–768. [Google Scholar]
- Barbaro, K.C.; Ferreira, M.L.; Cardoso, D.F.; Eickstedt, V.R.; Mota, I. Identification and neutralization of biological activities in the venoms of Loxosceles spiders. Braz. J. Med. Biol. Res. 1996, 29, 1491–1497. [Google Scholar]
- Cunha, R.B.; Barbaro, K.C.; Muramatsu, D.; Portaro, F.C.; Fontes, W.; de Sousa, M.V. Purification and characterization of loxnecrogin, a dermonecrotic toxin from Loxosceles gaucho brown spider venom. J. Protein Chem. 2003, 22, 135–146. [Google Scholar]
- Kalapothakis, E.; Chatzaki, M.; Goncalves-Dornelas, H.; de Castro, C.S.; Silvestre, F.G.; Laborne, F.V.; de Moura, J.F.; Veiga, S.S.; Chavez-Olortegui, C.; Granier, C.; Barbaro, K.C. The Loxtox protein family in Loxosceles intermedia (Mello-Leitao) venom. Toxicon 2007, 50, 938–946. [Google Scholar]
- Feitosa, L.; Gremski, W.; Veiga, S.S.; Elias, M.C.; Graner, E.; Mangili, O.C.; Brentani, R.R. Detection and characterization of metalloproteinases with gelatinolytic, fibronectinolytic and fibrinogenolytic activities in brown spider (Loxosceles intermedia) venom. Toxicon 1998, 36, 1039–1051. [Google Scholar]
- Veiga, S.S.; da Silveira, R.B.; Dreyfus, J.L.; Haoach, J.; Pereira, A.M.; Mangili, O.C.; Gremski, W. Identification of high molecular weight serine-proteases in Loxosceles intermedia (brown spider) venom. Toxicon 2000, 38, 825–839. [Google Scholar]
- Veiga, S.S.; Feitosa, L.; dos Santos, V.L.; de Souza, G.A.; Ribeiro, A.S.; Mangili, O.C.; Porcionatto, M.A.; Nader, H.B.; Dietrich, C.P.; Brentani, R.R.; Gremski, W. Effect of brown spider venom on basement membrane structures. Histochem. J. 2000, 32, 397–408. [Google Scholar]
- Veiga, S.S.; Zanetti, V.C.; Braz, A.; Mangili, O.C.; Gremski, W. Extracellular matrix molecules as targets for brown spider venom toxins. Braz. J. Med. Biol. Res. 2001, 34, 843–850. [Google Scholar]
- Veiga, S.S.; Zanetti, V.C.; Franco, C.R.; Trindade, E.S.; Porcionatto, M.A.; Mangili, O.C.; Gremski, W.; Dietrich, C.P.; Nader, H.B. In vivo and in vitro cytotoxicity of brown spider venom for blood vessel endothelial cells. Thromb. Res. 2001, 102, 229–237. [Google Scholar] [CrossRef] [PubMed]
- de Castro, C.S.; Silvestre, F.G.; Araujo, S.C.; Gabriel de, M.Y.; Mangili, O.C.; Cruz, I.; Chavez-Olortegui, C.; Kalapothakis, E. Identification and molecular cloning of insecticidal toxins from the venom of the brown spider Loxosceles intermedia. Toxicon 2004, 44, 273–280. [Google Scholar]
- dos Santos, L.D.; Dias, N.B.; Roberto, J.; Pinto, A.S.; Palma, M.S. Brown recluse spider venom: proteomic analysis and proposal of a putative mechanism of action. Protein Pept. Lett. 2009, 16, 933–943. [Google Scholar]
- Machado, L.F.; Laugesen, S.; Botelho, E.D.; Ricart, C.A.; Fontes, W.; Barbaro, K.C.; Roepstorff, P.; Sousa, M.V. Proteome analysis of brown spider venom: identification of loxnecrogin isoforms in Loxosceles gaucho venom. Proteomics 2005, 5, 2167–2176. [Google Scholar]
- Corzo, G.; Gilles, N.; Satake, H.; Villegas, E.; Dai, L.; Nakajima, T.; Haupt, J. Distinct primary structures of the major peptide toxins from the venom of the spider Macrothele gigas that bind to sites 3 and 4 in the sodium channel. FEBS Lett. 2003, 547, 43–50. [Google Scholar]
- Gremski, L.H.; da Silveira, R.B.; Chaim, O.M.; Probst, C.M.; Ferrer, V.P.; Nowatzki, J.; Weinschutz, H.C.; Madeira, H.M.; Gremski, W.; Nader, H.B.; Senff-Ribeiro, A.; Veiga, S.S. A novel expression profile of the Loxosceles intermedia spider venomous gland revealed by transcriptome analysis. Mol. Biosyst. 2010, 6, 2403–2416. [Google Scholar]
- Binford, G.J.; Cordes, M.H.; Wells, M.A. Sphingomyelinase D from venoms of Loxosceles spiders: evolutionary insights from cDNA sequences and gene structure. Toxicon 2005, 45, 547–560. [Google Scholar]
- Trevisan-Silva, D.; Gremski, L.H.; Chaim, O.M.; da Silveira, R.B.; Meissner, G.O.; Mangili, O.C.; Barbaro, K.C.; Gremski, W.; Veiga, S.S.; Senff-Ribeiro, A. Astacin-like metalloproteases are a gene family of toxins present in the venom of different species of the brown spider (genus Loxosceles). Biochimie 2010, 92, 21–32. [Google Scholar]
- Wood, D.L.; Miljenovic, T.; Cai, S.; Raven, R.J.; Kaas, Q.; Escoubas, P.; Herzig, V.; Wilson, D.; King, G.F. ArachnoServer: A database of protein toxins from spiders. BMC Genomics 2009, 10, 375. [Google Scholar]
- King, G.F.; Gentz, M.C.; Escoubas, P.; Nicholson, G.M. A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon 2008, 52, 264–276. [Google Scholar]
- Kalapothakis, E.; Araujo, S.C.; de Castro, C.S.; Mendes, T.M.; Gomez, M.V.; Mangili, O.C.; Gubert, I.C.; Chavez-Olortegui, C. Molecular cloning, expression and immunological properties of LiD1, a protein from the dermonecrotic family of Loxosceles intermedia spider venom. Toxicon 2002, 40, 1691–1699. [Google Scholar] [CrossRef] [PubMed]
- Tambourgi, D.V.; Fernandes-Pedrosa, M.F.; van den Berg, C.W.; Goncalves-de-Andrade, R.M.; Ferracini, M.; Paixao-Cavalcante, D.; Morgan, B.P.; Rushmere, N.K. Molecular cloning, expression, function and immunoreactivities of members of a gene family of sphingomyelinases from Loxosceles venom glands. Mol. Immunol. 2004, 41, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Appel, M.H.; da Silveira, R.B.; Chaim, O.M.; Paludo, K.S.; Silva, D.T.; Chaves, D.M.; da Silva, P.H.; Mangili, O.C.; Senff-Ribeiro, A.; Gremski, W.; Nader, H.B.; Veiga, S.S. Identification, cloning and functional characterization of a novel dermonecrotic toxin (phospholipase D) from brown spider (Loxosceles intermedia) venom. Biochim. Biophys. Acta 2008, 1780, 167–178. [Google Scholar] [PubMed]
- Chaim, O.M.; Sade, Y.B.; da Silveira, R.B.; Toma, L.; Kalapothakis, E.; Chavez-Olortegui, C.; Mangili, O.C.; Gremski, W.; von Dietrich, C.P.; Nader, H.B.; Veiga, S.S. Brown spider dermonecrotic toxin directly induces nephrotoxicity. Toxicol. Appl. Pharmacol. 2006, 211, 64–77. [Google Scholar]
- da Silveira, R.B.; Pigozzo, R.B.; Chaim, O.M.; Appel, M.H.; Dreyfuss, J.L.; Toma, L.; Mangili, O.C.; Gremski, W.; Dietrich, C.P.; Nader, H.B.; Veiga, S.S. Molecular cloning and functional characterization of two isoforms of dermonecrotic toxin from Loxosceles intermedia (brown spider) venom gland. Biochimie 2006, 88, 1241–1253. [Google Scholar]
- da Silveira, R.B.; Pigozzo, R.B.; Chaim, O.M.; Appel, M.H.; Silva, D.T.; Dreyfuss, J.L.; Toma, L.; Dietrich, C.P.; Nader, H.B.; Veiga, S.S.; Gremski, W. Two novel dermonecrotic toxins LiRecDT4 and LiRecDT5 from brown spider (Loxosceles intermedia) venom: From cloning to functional characterization. Biochimie 2007, 89, 289–300. [Google Scholar]
- Ribeiro, R.O.; Chaim, O.M.; da Silveira, R.B.; Gremski, L.H.; Sade, Y.B.; Paludo, K.S.; Senff-Ribeiro, A.; de Moura, J.; Chavez-Olortegui, C.; Gremski, W.; Nader, H.B.; Veiga, S.S. Biological and structural comparison of recombinant phospholipase D toxins from Loxosceles intermedia (brown spider) venom. Toxicon 2007, 50, 1162–1174. [Google Scholar]
- Ramos-Cerrillo, B.; Olvera, A.; Odell, G.V.; Zamudio, F.; Paniagua-Solis, J.; Alagon, A.; Stock, R.P. Genetic and enzymatic characterization of sphingomyelinase D isoforms from the North American fiddleback spiders Loxosceles boneti and Loxosceles reclusa. Toxicon 2004, 44, 507–514. [Google Scholar]
- Murakami, M.T.; Fernandes-Pedrosa, M.F.; de Andrade, S.A.; Gabdoulkhakov, A.; Betzel, C.; Tambourgi, D.V.; Arni, R.K. Structural insights into the catalytic mechanism of sphingomyelinases D and evolutionary relationship to glycerophosphodiester phosphodiesterases. Biochem. Biophys. Res. Commun. 2006, 342, 323–329. [Google Scholar]
- Lee, S.; Lynch, K.R. Brown recluse spider (Loxosceles reclusa) venom phospholipase D (PLD) generates lysophosphatidic acid (LPA). Biochem. J. 2005, 391, 317–323. [Google Scholar]
- Chaim, O.M.; da Silveira, R.B.; Trevisan-Silva, D.; Ferrer, V.P.; Sade, Y.B.; Bóia-Ferreira, M.; Gremski, L.H.; Gremski, W.; Senff-Ribeiro, A.; Takahashi, H.K.; Toledo, M.S.; Nader, H.B.; Veiga, S.S. Phospholipase-D activity and inflammatory response induced by brown spider dermonecrotic toxin: Endothelial cell membrane phospholipids as targets for toxicity. BBA Mol. Cell Biol. Lipids 2010, 1811, 84–96. [Google Scholar]
- Fernandes Pedrosa, M.F.; Junqueira de Azevedo I. de, L.; Goncalves-de-Andrade, R.M.; van den Berg, C.W.; Ramos, C.R.; Ho, P.L.; Tambourgi, D.V. Molecular cloning and expression of a functional dermonecrotic and haemolytic factor from Loxosceles laeta venom. Biochem. Biophys. Res. Commun. 2002, 298, 638–645. [Google Scholar]
- Tambourgi, D.V.; Magnoli, F.C.; Von Eickstedt, V.R.; Benedetti, Z.C.; Petricevich, V.L.; da Silva, W.D. Incorporation of a. 35-kilodalton purified protein from Loxosceles intermedia spider venom transforms human erythrocytes into activators of autologous complement alternative pathway. J. Immunol. 1995, 155, 4459–4466. [Google Scholar] [PubMed]
- Chaves-Moreira, D.; Chaim, O.M.; Sade, Y.B.; Paludo, K.S.; Gremski, L.H.; Donatti, L.; de Moura, J.; Mangili, O.C.; Gremski, W.; da Silveira, R.B.; Senff-Ribeiro, A.; Veiga, S.S. Identification of a direct hemolytic effect dependent on the catalytic activity induced by phospholipase-D (dermonecrotic toxin) from brown spider venom. J. Cell. Biochem. 2009, 107, 655–666. [Google Scholar]
- Kusma, J.; Chaim, O.M.; Wille, A.C.; Ferrer, V.P.; Sade, Y.B.; Donatti, L.; Gremski, W.; Mangili, O.C.; Veiga, S.S. Nephrotoxicity caused by brown spider venom phospholipase-D (dermonecrotic toxin) depends on catalytic activity. Biochimie 2008, 90, 1722–1736. [Google Scholar]
- Barbaro, K.C.; Sousa, M.V.; Morhy, L.; Eickstedt, V.R.; Mota, I. Compared chemical properties of dermonecrotic and lethal toxins from spiders of the genus Loxosceles (Araneae). J. Protein Chem. 1996, 15, 337–343. [Google Scholar]
- van Meeteren, L.A.; Frederiks, F.; Giepmans, B.N.; Pedrosa, M.F.; Billington, S.J.; Jost, B.H.; Tambourgi, D.V.; Moolenaar, W.H. Spider and bacterial sphingomyelinases D target cellular lysophosphatidic acid receptors by hydrolyzing lysophosphatidylcholine. J. Biol. Chem. 2004, 279, 10833–10836. [Google Scholar]
- Dragulev, B.; Bao, Y.; Ramos-Cerrillo, B.; Vazquez, H.; Olvera, A.; Stock, R.; Algaron, A.; Fox, J.W. Upregulation of IL-6, IL-8, CXCL1, and CXCL2 dominates gene expression in human fibroblast cells exposed to Loxosceles reclusa sphingomyelinase D: Insights into spider venom dermonecrosis. J. Invest. Dermatol. 2007, 127, 1264–1266. [Google Scholar] [PubMed]
- Barrett, S.M.; Romine-Jenkins, M.; Blick, K.E. Passive hemagglutination inhibition test for diagnosis of brown recluse spider bite envenomation. Clin. Chem. 1993, 39, 2104–2107. [Google Scholar]
- Fernandes-Pedrosa, F.; Junqueira-de-Azevedo, L.; Goncalves-de-Andrade, R.M.; Kobashi, L.S.; Almeida, D.D.; Ho, P.L.; Tambourgi, D.V. Transcriptome analysis of Loxosceles laeta (Araneae, Sicariidae) spider venomous gland using expressed sequence tags. BMC Genomics 2008, 9, 279. [Google Scholar]
- da Silveira, R.B.; Wille, A.C.; Chaim, O.M.; Appel, M.H.; Silva, D.T.; Franco, C.R.; Toma, L.; Mangili, O.C.; Gremski, W.; Dietrich, C.P.; Nader, H.B.; Veiga, S.S. Identification, cloning, expression and functional characterization of an astacin-like metalloprotease toxin from Loxosceles intermedia (brown spider) venom. Biochem. J. 2007, 406, 355–363. [Google Scholar] [CrossRef] [PubMed]
- da Silveira, R.B.; dos Santos Filho, J.F.; Mangili, O.C.; Veiga, S.S.; Gremski, W.; Nader, H.B.; von Dietrich, C.P. Identification of proteases in the extract of venom glands from brown spiders. Toxicon 2002, 40, 815–822. [Google Scholar]
- Stocker, W.; Grams, F.; Baumann, U.; Reinemer, P.; Gomis-Ruth, F.X.; McKay, D.B.; Bode, W. The metzincins--topological and sequential relations between the astacins, adamalysins, serralysins, and matrixins (collagenases) define a superfamily of zinc-peptidas. Protein Sci. 1995, 4, 823–840. [Google Scholar]
- Wright, R.P.; Elgert, K.D.; Campbell, B.J.; Barrett, J.T. Hyaluronidase and esterase activities of the venom of the poisonous brown recluse spider. Arch. Biochem. Biophys. 1973, 159, 415–426. [Google Scholar]
- Bommer, U.A.; Thiele, B.J. The translationally controlled tumour protein (TCTP). Int. J. Biochem. Cell Biol. 2004, 36, 379–385. [Google Scholar]
- Marsh, N.; Williams, V. Practical applications of snake venom toxins in haemostasis. Toxicon 2005, 45, 1171–1181. [Google Scholar]
- Koh, D.C.; Armugam, A.; Jeyaseelan, K. Snake venom components and their applications in biomedicine. Cell Mol. Life Sci. 2006, 63, 3030–3041. [Google Scholar]
- Bailey, P.; Wilce, J. Venom as a source of useful biologically active molecules. Emerg. Med. (Fremantle) 2001, 13, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Schmidtko, A.; Lotsch, J.; Freynhagen, R.; Geisslinger, G. Ziconotide for treatment of severe chronic pain. Lancet 2010, 375, 1569–1577. [Google Scholar]
- Ramu, Y.; Xu, Y.; Lu, Z. Engineered specific and high-affinity inhibitor for a subtype of inward-rectifier K+ channels. Proc. Natl. Acad. Sci. USA 2008, 105, 10774–10778. [Google Scholar]
- Gedulin, B.R.; Smith, P.; Prickett, K.S.; Tryon, M.; Barnhill, S.; Reynolds, J.; Nielsen, L.L.; Parkes, D.G.; Young, A.A. Dose-response for glycaemic and metabolic changes. 28 days after single injection of long-acting release exenatide in diabetic fatty Zucker rats. Diabetologia 2005, 48, 1380–1385. [Google Scholar] [CrossRef] [PubMed]
- Heine, R.J.; Van Gaal, L.F.; Johns, D.; Mihm, M.J.; Widel, M.H.; Brodows, R.G. Exenatide versus insulin glargine in patients with suboptimally controlled type. 2 diabetes: A randomized trial. Ann. Intern. Med. 2005, 143, 559–569. [Google Scholar] [PubMed]
- Chuang, R.S.; Jaffe, H.; Cribbs, L.; Perez-Reyes, E.; Swartz, K.J. Inhibition of T-type voltage-gated calcium channels by a new scorpion toxin. Nat. Neurosci. 1998, 1, 668–674. [Google Scholar]
- Bagdany, M.; Batista, C.V.; Valdez-Cruz, N.A.; Somodi, S.; Rodriguez de la Vega, R.C.; Licea, A.F.; Varga, Z.; Gaspar, R.; Possani, L.D.; Panyi, G. Anuroctoxin, a new scorpion toxin of the alpha-KTx. 6 subfamily, is highly selective for Kv1.3 over IKCa1 ion channels of human T lymphocytes. Mol. Pharmacol. 2005, 67, 1034–1044. [Google Scholar] [CrossRef] [PubMed]
- Gurevitz, M.; Karbat, I.; Cohen, L.; Ilan, N.; Kahn, R.; Turkov, M.; Stankiewicz, M.; Stuhmer, W.; Dong, K.; Gordon, D. The insecticidal potential of scorpion beta-toxins. Toxicon 2007, 49, 473–489. [Google Scholar]
- Diochot, S.; Lazdunski, M. Sea anemone toxins affecting potassium channels. Prog. Mol. Subcell. Biol. 2009, 46, 99–122. [Google Scholar]
- Mirshafiey, A. Venom therapy in multiple sclerosis. Neuropharmacology 2007, 53, 353–361. [Google Scholar]
- Norton, R.S.; Pennington, M.W.; Wulff, H. Potassium channel blockade by the sea anemone toxin ShK for the treatment of multiple sclerosis and other autoimmune diseases. Curr. Med. Chem. 2004, 11, 3041–3052. [Google Scholar]
- Fletcher, J.I.; Smith, R.; O'Donoghue, S.I.; Nilges, M.; Connor, M.; Howden, M.E.; Christie, M.J.; King, G.F. The structure of a novel insecticidal neurotoxin, omega-atracotoxin-HV1, from the venom of an Australian funnel web spider. Nat. Struct. Biol. 1997, 4, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Villanova, F.E.; Andrade, E.; Leal, E.; Andrade, P.M.; Borra, R.C.; Troncone, L.R.; Magalhaes, L.; Leite, K.R.; Paranhos, M.; Claro, J.; Srougi, M. Erection induced by Tx2-6 toxin of Phoneutria nigriventer spider: expression profile of genes in the nitric oxide pathway of penile tissue of mice. Toxicon 2009, 54, 793–801. [Google Scholar]
- Andrade, E.; Villanova, F.; Borra, P.; Leite, K.; Troncone, L.; Cortez, I.; Messina, L.; Paranhos, M.; Claro, J.; Srougi, M. Penile erection induced in vivo by a purified toxin from the Brazilian spider Phoneutria nigriventer. BJU Int. 2008, 102, 835–837. [Google Scholar] [PubMed]
- Haeberli, S.; Kuhn-Nentwig, L.; Schaller, J.; Nentwig, W. Characterisation of antibacterial activity of peptides isolated from the venom of the spider Cupiennius salei (Araneae: Ctenidae). Toxicon 2000, 38, 373–380. [Google Scholar]
- Senff-Ribeiro, A.; Henrique da Silva, P.; Chaim, O.M.; Gremski, L.H.; Paludo, K.S.; Bertoni da Silveira, R.; Gremski, W.; Mangili, O.C.; Veiga, S.S. Biotechnological applications of brown spider (Loxosceles genus) venom toxins. Biotechnol. Adv. 2008, 26, 210–218. [Google Scholar]
- Veiga, S.S. Federal University of Paraná, Paraná, Brazil, Personal communication, 2011.
- Binford, G.J.; Wells, M.A. The phylogenetic distribution of sphingomyelinase D activity in venoms of Haplogyne spiders. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2003, 135, 25–33. [Google Scholar]
- Tambourgi, D.V.; Goncalves-de-Andrade, R.M.; van den Berg, C.W. Loxoscelism: From basic research to the proposal of new therapies. Toxicon 2010, 56, 1113–1119. [Google Scholar]
- Barbaro, K.C.; Eickstedt, V.R.; Mota, I. Antigenic cross-reactivity of venoms from medically important Loxosceles (Araneae) species in Brazil. Toxicon 1994, 32, 113–120. [Google Scholar]
- Mota, I.; Barbaro, K.C. Biological and biochemical properties of venoms from medically important Loxosceles (Araneae) species in Brazil. Toxin Rev. 1995, 14, 401–421. [Google Scholar]
- Luciano, M.N.; da Silva, P.H.; Chaim, O.M.; dos Santos, V.L.; Franco, C.R.; Soares, M.F.; Zanata, S.M.; Mangili, O.C.; Gremski, W.; Veiga, S.S. Experimental evidence for a direct cytotoxicity of Loxosceles intermedia (brown spider) venom in renal tissue. J. Histochem. Cytochem. 2004, 52, 455–467. [Google Scholar]
- Paixao-Cavalcante, D.; van den Berg, C.W.; de Freitas Fernandes-Pedrosa, M.; Goncalves de Andrade, R.M.; Tambourgi, D.V. Role of matrix metalloproteinases in HaCaT keratinocytes apoptosis induced by loxosceles venom sphingomyelinase D. J. Invest. Dermatol. 2006, 126, 61–68. [Google Scholar]
- McGlasson, D.L.; Green, J.A.; Stoecker, W.V.; Babcock, J.L.; Calcara, D.A. Duration of Loxosceles reclusa venom detection by ELISA from swabs. Clin. Lab. Sci. 2009, 22, 216–222. [Google Scholar]
- Vetter, R.S. Arachnids misidentified as brown recluse spiders by medical personnel and other authorities in North America. Toxicon 2009, 54, 545–547. [Google Scholar]
- Reitz, M. Diagnosis of brown recluse spider bites is overused. Am. Fam. Physician 2007, 76, 943–944. [Google Scholar]
- Pauli, I.; Minozzo, J.C.; da Silva, P.H.; Chaim, O.M.; Veiga, S.S. Analysis of therapeutic benefits of antivenin at different time intervals after experimental envenomation in rabbits by venom of the Brown spider (Loxosceles intermedia). Toxicon 2009, 53, 660–671. [Google Scholar]
- Pauli, I.; Puka, J.; Gubert, I.C.; Minozzo, J.C. The efficacy of antivenom in loxoscelism treatment. Toxicon 2006, 48, 123–137. [Google Scholar]
- Dias-Lopes, C.; Guimaraes, G.; Felicori, L.; Fernandes, P.; Emery, L.; Kalapothakis, E.; Nguyen, C.; Molina, F.; Granier, C.; Chavez-Olortegui, C. A protective immune response against lethal, dermonecrotic and hemorrhagic effects of Loxosceles intermedia venom elicited by a. 27-residue peptide. Toxicon 2010, 55, 481–487. [Google Scholar]
- Felicori, L.; Fernandes, P.B.; Giusta, M.S.; Duarte, C.G.; Kalapothakis, E.; Nguyen, C.; Molina, F.; Granier, C.; Chavez-Olortegui, C. An in. vivo protective response against toxic effects of the dermonecrotic protein from Loxosceles intermedia spider venom elicited by synthetic epitopes. Vaccine 2009, 27, 4201–4208. [Google Scholar] [PubMed]
- Gomez, H.F.; Krywko, D.M.; Stoecker, W.V. A new assay for the detection of Loxosceles species (brown recluse) spider venom. Ann. Emerg. Med. 2002, 39, 469–474. [Google Scholar]
- Krywko, D.M.; Gomez, H.F. Detection of Loxosceles species venom in dermal lesions: A comparison of 4 venom recovery methods. Ann. Emerg. Med. 2002, 39, 475–480. [Google Scholar]
- McDermott, M.; Wakelam, M.J.; Morris, A.J. Phospholipase D. Biochem. Cell. Biol. 2004, 82, 225–253. [Google Scholar]
- Gomez-Cambronero, J. New concepts in phospholipase D signaling in inflammation and cancer. Sci. World J. 2010, 10, 1356–1369. [Google Scholar]
- Roth, M.G. Molecular mechanisms of PLD function in membrane traffic. Traffic 2008, 9, 1233–1239. [Google Scholar]
- Huwiler, A.; Kolter, T.; Pfeilschifter, J.; Sandhoff, K. Physiology and pathophysiology of sphingolipid metabolism and signaling. Biochim. Biophys. Acta 2000, 1485, 63–99. [Google Scholar] [PubMed]
- Mitsutake, S.; Igarashi, Y. Transbilayer movement of ceramide in the plasma membrane of live cells. Biochem. Biophys. Res. Commun. 2007, 359, 622–627. [Google Scholar]
- Rodrigues, R.S.; Izidoro, L.F.; de Oliveira, R.J., Jr.; Sampaio, S.V.; Soares, A.M.; Rodrigues, V.M. Snake venom phospholipases A2: A new class of antitumor agents. Protein Pept. Lett. 2009, 16, 894–898. [Google Scholar]
- Su, W.; Chen, Q.; Frohman, M.A. Targeting phospholipase D with small-molecule inhibitors as a potential therapeutic approach for cancer metastasis. Future Oncol. 2009, 5, 1477–1486. [Google Scholar]
- Majd, S.; Yusko, E.C.; MacBriar, A.D.; Yang, J.; Mayer, M. Gramicidin pores report the activity of membrane-active enzymes. J. Am. Chem. Soc. 2009, 131, 16119–16126. [Google Scholar]
- Ramu, Y.; Xu, Y.; Lu, Z. Enzymatic activation of voltage-gated potassium channels. Nature 2006, 442, 696–699. [Google Scholar]
- Menzel, E.J.; Farr, C. Hyaluronidase and its substrate hyaluronan: Biochemistry, biological activities and therapeutic uses. Cancer Lett. 1998, 131, 3–11. [Google Scholar]
- Cramer, J.A.; Bailey, L.C.; Bailey, C.A.; Miller, R.T. Kinetic and mechanistic studies with bovine testicular hyaluronidase. Biochim. Biophys. Acta 1994, 1200, 315–321. [Google Scholar] [PubMed]
- Girish, K.S.; Kemparaju, K. The magic glue hyaluronan and its eraser hyaluronidase: A biological overview. Life Sci. 2007, 80, 1921–1943. [Google Scholar]
- Kemparaju, K.; Girish, K.S. Snake venom hyaluronidase: a therapeutic target. Cell Biochem. Funct. 2006, 24, 7–12. [Google Scholar]
- Magalhaes, M.R.; da Silva, N.J., Jr.; Ulhoa, C.J. A hyaluronidase from Potamotrygon motoro (freshwater stingrays) venom: Isolation and characterization. Toxicon 2008, 51, 1060–1067. [Google Scholar]
- Girish, K.S.; Kemparaju, K. A low molecular weight isoform of hyaluronidase: purification from Indian cobra (Naja naja) venom and partial characterization. Biochemistry (Mosc) 2005, 70, 708–712. [Google Scholar] [CrossRef] [PubMed]
- Markovic-Housley, Z.; Miglierini, G.; Soldatova, L.; Rizkallah, P.J.; Muller, U.; Schirmer, T. Crystal structure of hyaluronidase, a major allergen of bee venom. Structure 2000, 8, 1025–1035. [Google Scholar]
- Skov, L.K.; Seppala, U.; Coen, J.J.; Crickmore, N.; King, T.P.; Monsalve, R.; Kastrup, J.S.; Spangfort, M.D.; Gajhede, M. Structure of recombinant Ves v. 2 at 2.0 Angstrom resolution: structural analysis of an allergenic hyaluronidase from wasp venom. Acta Crystallogr. D Biol. Crystallogr. 2006, 62, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Tan, N.H.; Ponnudurai, G. Comparative study of the enzymatic, hemorrhagic, procoagulant and anticoagulant activities of some animal venoms. Comp. Biochem. Physiol. C 1992, 103, 299–302. [Google Scholar] [PubMed]
- Kaiser, E. Trypsin and hyaluronidase inhibitor of human serum; the inhibition of the proteolytic and hyaluronic acid cleavage enzymes of snake and spider venoms by human serum. Biochem. J. 1953, 324, 344–350. [Google Scholar]
- Nagaraju, S.; Devaraja, S.; Kemparaju, K. Purification and properties of hyaluronidase from Hippasa partita (funnel web spider) venom gland extract. Toxicon 2007, 50, 383–393. [Google Scholar]
- Rash, L.D.; Hodgson, W.C. Pharmacology and biochemistry of spider venoms. Toxicon 2002, 40, 225–254. [Google Scholar]
- Kuhn-Nentwig, L.; Schaller, J.; Nentwig, W. Biochemistry, toxicology and ecology of the venom of the spider Cupiennius salei (Ctenidae). Toxicon 2004, 43, 543–553. [Google Scholar]
- Nagaraju, S.; Mahadeswaraswamy, Y.H.; Girish, K.S.; Kemparaju, K. Venom from spiders of the genus Hippasa: Biochemical and pharmacological studies. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2006, 144, 1–9. [Google Scholar]
- Rocha-e-Silva, T.A.A.; Sutti, R.; Hyslop, S. Milking and partial characterization of venom from the Brazilian spider Vitalius dubius (Theraphosidae). Toxicon 2009, 53, 153–161. [Google Scholar]
- Zobel-Thropp, P.A.; Bodner, M.R.; Binford, G.J. Comparative analyses of venoms from American and African Sicarius spiders that differ in sphingomyelinase D activity. Toxicon 2010, 55, 1274–1282. [Google Scholar]
- Goolsby, T.V.; Lombardo, F.A. Extravasation of Chemotherapeutic Agents: Prevention and Treatment. Semin.Oncol. 2006, 33, 139–143. [Google Scholar]
- Dunn, A.L.; Heavner, J.E.; Racz, G.; Day, M. Hyaluronidase: A review of approved formulations, indications and off-label use in chronic pain management. Expert Opin. Biol. Ther. 2010, 10, 127–131. [Google Scholar]
- Muchmore, D.B.; Vaughn, D.E. Review of the mechanism of action and clinical efficacy of recombinant human hyaluronidase coadministration with current prandial insulin formulations. J. Diabetes Sci. Technol. 2010, 4, 419–428. [Google Scholar]
- Etesse, B.; Beaudroit, L.; Deleuze, M.; Nouvellon, E.; Ripart, J. Hyaluronidase: Here we go again. Ann. Fr. Anesth. Reanim. 2009, 28, 658–665. [Google Scholar]
- Misbah, S.; Sturzenegger, M.H.; Borte, M.; Shapiro, R.S.; Wasserman, R.L.; Berger, M.; Ochs, H.D. Subcutaneous immunoglobulin: opportunities and outlook. Clin. Exp. Immunol. 2009, 158, 51–59. [Google Scholar]
- Lokeshwar, V.B.; Selzer, M.G. Hyalurondiase: Both a tumor promoter and suppressor. Semin. Cancer Biol. 2008, 18, 281–287. [Google Scholar]
- Barla, F.; Higashijima, H.; Funai, S.; Sugimoto, K.; Harada, N.; Yamaji, R.; Fujita, T.; Nakano, Y.; Inui, H. Inhibitive effects of alkyl gallates on hyaluronidase and collagenase. Biosci. Biotechnol. Biochem. 2009, 73, 2335–2337. [Google Scholar]
- Shuster, S.; Frost, G.I.; Csoka, A.B.; Formby, B.; Stern, R. Hyaluronidase reduces human breast cancer xenografts in SCID mice. Int. J. Cancer 2002, 102, 192–197. [Google Scholar]
- Botzki, A.; Rigden, D.J.; Braun, S.; Nukui, M.; Salmen, S.; Hoechstetter, J.; Bernhardt, G.; Dove, S.; Jedrzejas, M.J.; Buschauer, A. L-Ascorbic acid. 6-hexadecanoate, a potent hyaluronidase inhibitor. X-ray structure and molecular modeling of enzyme-inhibitor complexes. J. Biol. Chem. 2004, 279, 45990–45997. [Google Scholar]
- Calvete, J.J. Venomics: Digging into the evolution of venomous systems and learning to twist nature to fight pathology. J. Proteomics 2009, 72, 121–126. [Google Scholar]
- Escoubas, P.; King, G.F. Venomics as a drug discovery platform. Expert Rev. Proteomics 2009, 6, 221–224. [Google Scholar]
- Gross, B.; Gaestel, M.; Bohm, H.; Bielka, H. cDNA sequence coding for a translationally controlled human tumor. Protein Nucleic Acids Res. 1989, 17, 8367. [Google Scholar]
- MacDonald, S.M.; Rafnar, T.; Langdon, J.; Lichtenstein, L.M. Molecular identification of an IgE-dependent histamine-releasing factor. Science 1995, 269, 688–690. [Google Scholar]
- Choi, K.W.; Hsu, Y.C. To cease or to proliferate: New insights into TCTP function from a Drosophila study. Cell Adh. Migr. 2007, 1, 129–130. [Google Scholar]
- Sun, J.; Wu, Y.; Wang, J.; Ma, F.; Liu, X.; Li, Q. Novel translationally controlled tumor protein homologue in the buccal gland secretion of Lampetra japonica. Biochimie 2008, 90, 1760–1768. [Google Scholar]
- Gachet, Y.; Tournier, S.; Lee, M.; Lazaris-Karatzas, A.; Poulton, T.; Bommer, U.A. The growth-related, translationally controlled protein P23 has properties of a tubulin binding protein and associates transiently with microtubules during the cell cycle. J. Cell. Sci. 1999, 112, 1257–1271. [Google Scholar]
- Bazile, F.; Pascal, A.; Arnal, I.; Le Clainche, C.; Chesnel, F.; Kubiak, J.Z. Complex relationship between TCTP, microtubules and actin microfilaments regulates cell shape in normal and cancer cells. Carcinogenesis 2009, 30, 555–565. [Google Scholar] [Green Version]
- Yarm, F.R. Plk phosphorylation regulates the microtubule-stabilizing protein TCTP. Mol. Cell. Biol. 2002, 22, 6209–6221. [Google Scholar]
- Cans, C.; Passer, B.J.; Shalak, V.; Nancy-Portebois, V.; Crible, V.; Amzallag, N.; Allanic, D.; Tufino, R.; Argentini, M.; Moras, D.; Fiucci, G.; Goud, B.; Mirande, M.; Amson, R.; Telerman, A. Translationally controlled tumor protein acts as a guanine nucleotide dissociation inhibitor on the translation elongation factor eEF1A. Pro. Natl. Acad. Sci. USA 2003, 100, 13892–13897. [Google Scholar]
- Liu, H.; Peng, H.W.; Cheng, Y.S.; Yuan, H.S.; Yang-Yen, H.F. Stabilization and enhancement of the antiapoptotic activity of mcl-1 by TCTP. Mol. Cell. Biol. 2005, 25, 3117–3126. [Google Scholar]
- Li, F.; Zhang, D.; Fujise, K. Characterization of fortilin, a novel antiapoptotic. ProteinJ. Biol. Chem. 2001, 276, 47542–47549. [Google Scholar]
- Amzallag, N.; Passer, B.J.; Allanic, D.; Segura, E.; Thery, C.; Goud, B.; Amson, R.; Telerman, A. TSAP6 facilitates the secretion of translationally controlled tumor protein-histamine-releasing factor via a nonclassical pathway. J. Biol. Chem. 2004, 279, 46104–46112. [Google Scholar]
- Jung, J.; Kim, M.; Kim, M.J.; Kim, J.; Moon, J.; Lim, J.S.; Kim, M.; Lee, K. Translationally controlled tumor protein interacts with the third cytoplasmic domain of Na,K-ATPase alpha subunit and inhibits the pump activity in HeLa cells. J. Biol. Chem. 2004, 279, 49868–49875. [Google Scholar]
- Yang, Y.; Yang, F.; Xiong, Z.; Yan, Y.; Wang, X.; Nishino, M.; Mirkovic, D.; Nguyen, J.; Wang, H.; Yang, X.F. An N-terminal region of translationally controlled tumor protein is required for its antiapoptotic activity. Oncogene 2005, 24, 4778–4788. [Google Scholar]
- Burgess, A.; Labbe, J.C.; Vigneron, S.; Bonneaud, N.; Strub, J.M.; Van Dorsselaer, A.; Lorca, T.; Castro, A. Chfr interacts and colocalizes with TCTP to the mitotic spindle. Oncogene 2008, 27, 5554–5566. [Google Scholar]
- Chen, S.H.; Wu, P.S.; Chou, C.H.; Yan, Y.T.; Liu, H.; Weng, S.Y.; Yang-Yen, H.F. A knockout mouse approach reveals that TCTP functions as an essential factor for cell proliferation and survival in a tissue- or cell type-specific manner. Mol. Biol. Cell 2007, 18, 2525–2532. [Google Scholar]
- Hsu, Y.; Chern, J.J.; Cai, Y.; Liu, M.; Choi, K.W. Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature 2007, 445, 785–788. [Google Scholar]
- Bheekha-Escura, R.; MacGlashan, D.W., Jr.; Langdon, J.M.; MacDonald, S.M. Human recombinant histamine-releasing factor activates human eosinophils and the eosinophilic cell line, AML14-3D10. Blood 2000, 96, 2191. [Google Scholar]
- Kang, H.S.; Lee, M.J.; Song, H.; Han, S.H.; Kim, Y. M.; Im, J.Y.; Choi, I. Molecular Identification of IgE-Dependent Histamine-Releasing Factor as a B Cell Growth Factor. 1. J. Immunol. 2001, 166, 6545–6554. [Google Scholar] [PubMed]
- Hinojosa-Moya, J.; Xoconostle-Cazares, B.; Piedra-Ibarra, E.; Mendez-Tenorio, A.; Lucas, W.J.; Ruiz-Medrano, R. Phylogenetic and structural analysis of translationally controlled tumor proteins. J. Mol. Evol. 2008, 66, 472–483. [Google Scholar]
- Mulenga, A.; Azad, A.F. The molecular and biological analysis of ixodid ticks histamine release factors. Exp. Appl. Acarology 2005, 37, 215–229. [Google Scholar]
- Rattmann, Y.D.; Pereira, C.R.; Cury, Y.; Gremski, W.; Marques, M.C.A.; da Silva-Santos, J.E. Vascular permeability and vasodilation induced by the Loxosceles intermedia venom in rats: Involvement of mast cell degranulation, histamine and. 5-HT receptors. Toxicon 2008, 51, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Rash, L.D.; King, R.G.; Hodgson, W.C. Evidence that histamine is the principal pharmacological component of venom from an Australian wolf spider (Lycosa godeffroyi). Toxicon 1998, 36, 367–375. [Google Scholar]
- Paludo, K.S.; Biscaia, S.M.; Chaim, O.M.; Otuki, M.F.; Naliwaiko, K.; Dombrowski, P.A.; Franco, C.R.; Veiga, S.S. Inflammatory events induced by brown spider venom and its recombinant dermonecrotic toxin: A pharmacological investigation. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2009, 149, 323–333. [Google Scholar]
- Weisel-Eichler, A.; Libersat, F. Venom effects on monoaminergic systems. J. Comp. Physiol. A Neuroethology Sens. Neural Behav. Physiol. 2004, 190, 683–690. [Google Scholar]
- Rattmann, Y.D.; Pereira, C.R.; Cury, Y.; Gremski, W.; Marques, M.C.; da Silva-Santos, J.E. Vascular permeability and vasodilation induced by the Loxosceles intermedia venom in rats: involvement of mast cell degranulation, histamine and. 5-HT receptors. Toxicon 2008, 51, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Gnanasekar, M.; Rao, K.V.; Chen, L.; Narayanan, R.B.; Geetha, M.; Scott, A.L.; Ramaswamy, K.; Kaliraj, P. Molecular characterization of a calcium binding translationally controlled tumor protein homologue from the filarial parasites Brugia malayi and Wuchereria bancrofti. Mol. Biochem. Parasitol. 2002, 121, 107–118. [Google Scholar]
- MacDonald, S.M.; Bhisutthibhan, J.; Shapiro, T.A.; Rogerson, S.J.; Taylor, T.E.; Tembo, M.; Langdon, J.M.; Meshnick, S.R. Immune mimicry in malaria: Plasmodium falciparum secretes a functional histamine-releasing factor homolog in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2001, 98, 10829–10832. [Google Scholar]
- Rao, K.V.; Chen, L.; Gnanasekar, M.; Ramaswamy, K. Cloning and characterization of a calcium-binding, histamine-releasing protein from Schistosoma mansoni. J. Biol. Chem. 2002, 277, 31207–31213. [Google Scholar]
- Efferth, T. Antiplasmodial and antitumor activity of artemisinin—From bench to bedside. Planta Med. 2007, 73, 299. [Google Scholar]
- Susini, L.; Besse, S.; Duflaut, D.; Lespagnol, A.; Beekman, C.; Fiucci, G.; Atkinson, A.R.; Busso, D.; Poussin, P.; Marine, J.C.; Martinou, J.C.; Cavarelli, J.; Moras, D.; Amson, R.; Telerman, A. TCTP protects from apoptotic cell death by antagonizing bax function. Cell Death Differ. 2008, 15, 1211–1220. [Google Scholar]
- Gnanasekar, M.; Thirugnanam, S.; Zheng, G.; Chen, A.; Ramaswamy, K. Gene silencing of translationally controlled tumor protein (TCTP) by siRNA inhibits cell growth and induces apoptosis of human prostate cancer cells. Int. J. Oncol. 2009, 34, 1241–1246. [Google Scholar]
- Telerman, A.; Amson, R. The molecular programme of tumour reversion: the steps beyond malignant transformation. Nat. Rev. Cancer 2009, 9, 206–216. [Google Scholar]
- Tuynder, M.; Fiucci, G.; Prieur, S.; Lespagnol, A.; Geant, A.; Beaucourt, S.; Duflaut, D.; Besse, S.; Susini, L.; Cavarelli, J.; Moras, D.; Amson, R.; Telerman, A. Translationally controlled tumor protein is a target of tumor reversion. Proc. Natl. Acad. Sci. USA 2004, 101, 15364–15369. [Google Scholar]
- Slaby, O.; Sobkova, K.; Svoboda, M.; Garajova, I.; Fabian, P.; Hrstka, R.; Nenutil, R.; Sachlova, M.; Kocakova, I.; Michalek, J.; Smerdova, T.; Knoflickova, D.; Vyzula, R. Significant overexpression of Hsp110 gene during colorectal cancer progression. Oncol. Rep. 2009, 21, 1235–1241. [Google Scholar]
- Ma, Q.; Geng, Y.; Xu, W.; Wu, Y.; He, F.; Shu, W.; Huang, M.; Du, H.; Li, M. The Role of Translationally Controlled Tumor Protein in Tumor Growth and Metastasis of Colon Adenocarcinoma Cells. J. Proteome. Res. 2009, 9, 40–49. [Google Scholar]
- Tuynder, M.; Susini, L.; Prieur, S.; Besse, S.; Fiucci, G.; Amson, R.; Telerman, A. Biological models and genes of tumor reversion: Cellular reprogramming through tpt1/TCTP and SIAH-1. Proc. Natl. Acad. Sci. USA 2002, 99, 14976–14981. [Google Scholar]
- Efferth, T. Mechanistic perspectives for. 1,2,4-trioxanes in anti-cancer therapy. Drug Resist. Updat. 2005, 8, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Koo, K.H.; Kim, Y.H.; Sohn, J.; Park, Y.G. Identification of potential lung cancer biomarkers using an in vitro carcinogenesis model. Exp. Mol. Med. 2008, 40, 709–720. [Google Scholar]
- van de Sande, W.W.; Janse, D.J.; Hira, V.; Goedhart, H.; van der Zee, R.; Ahmed, A.O.; Ott, A.; Verbrugh, H.; van Belkum, A. Translationally controlled tumor protein from Madurella mycetomatis, a marker for tumorous mycetoma progression. J. Immunol. 2006, 177, 1997–2005. [Google Scholar]
- Zhu, W.L.; Cheng, H.X.; Han, N.; Liu, D.L.; Zhu, W.X.; Fan, B.L.; Duan, F.L. Messenger RNA expression of translationally controlled tumor protein (TCTP) in liver regeneration and cancer. Anticancer Res. 2008, 28, 1575–1580. [Google Scholar]
- Rinnerthaler, M.; Jarolim, S.; Heeren, G.; Palle, E.; Perju, S.; Klinger, H.; Bogengruber, E.; Madeo, F.; Braun, R.J.; Breitenbach-Koller, L.; Breitenbach, M.; Laun, P. MMI1 (YKL056c, TMA19), the yeast orthologue of the translationally controlled tumor protein (TCTP) has apoptotic functions and interacts with both microtubules and mitochondria. Biochim. Biophys. Acta 2006, 1757, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, V.C.; da Silveira, R.B.; Dreyfuss, J.L.; Haoach, J.; Mangili, O.C.; Veiga, S.S.; Gremski, W. Morphological and biochemical evidence of blood vessel damage and fibrinogenolysis triggered by brown spider venom. Blood Coagul. Fibrinolysis 2002, 13, 135–148. [Google Scholar]
- Gomis-Rüth, F. Structural aspects of the metzincin clan of metalloendopeptidases. Mol. Biotechnol. 2003, 24, 157–202. [Google Scholar]
- Sterchi, E.E. Special issue: Metzincin metalloproteinases. Mol. Aspects Med. 2008, 29, 255–257. [Google Scholar]
- Becker-Pauly, C.; Bruns, B.C.; Damm, O.; Schutte, A.; Hammouti, K.; Burmester, T.; Stocker, W. News from an ancient world: two novel astacin metalloproteases from the horseshoe crab. J. Mol. Biol. 2009, 385, 236–248. [Google Scholar]
- Sarras, M.P., Jr. BMP-1 and the astacin family of metalloproteinases: A potential link between the extracellular matrix, growth factors and pattern formation. Bioessays 1996, 18, 439–442. [Google Scholar] [CrossRef] [PubMed]
- Mohrlen, F.; Hutter, H.; Zwilling, R. The astacin protein family in Caenorhabditis elegans. Eur. J. Biochem. 2003, 270, 4909–4920. [Google Scholar]
- Bode, W.; Gomis-Ruth, F.X.; Stockler, W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the 'metzincins'. FEBS Lett. 1993, 331, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Bond, J.S.; Beynon, R.J. The astacin family of metalloendopeptidases. Protein Sci. 1995, 4, 1247–1261. [Google Scholar]
- Stocker, W.; Zwilling, R. Astacin. Methods Enzymol. 1995, 248, 305–325. [Google Scholar]
- Stocker, W.; Bode, W. Structural features of a superfamily of zinc-endopeptidases: The metzincins. Curr. Opin. Struct. Biol. 1995, 5, 383–390. [Google Scholar]
- Dumermuth, E.; Sterchi, E.E.; Jiang, W.P.; Wolz, R.L.; Bond, J.S.; Flannery, A.V.; Beynon, R.J. The astacin family of metalloendopeptidases. J. Biol. Chem. 1991, 266, 21381–21385. [Google Scholar]
- Sharma, V.K.; Teoh, H.L.; Wong, L.Y.; Su, J.; Ong, B.K.; Chan, B.P. Recanalization therapies in acute ischemic stroke: pharmacological agents, devices, and combinations. Stroke Res. Treat. 2010, in press. [Google Scholar]
- Gao, F.; Kiesewetter, D.; Chang, L.; Ma, K.; Rapoport, S.I.; Igarashi, M. Whole-body synthesis secretion of docosahexaenoic acid from circulating eicosapentaenoic acid in unanesthetized rats. J. Lipid Res. 2009, 50, 2463–2470. [Google Scholar]
- Rash, L.D.; Hodgson, W.C. Pharmacology and biochemistry of spider venoms. Toxicon 2002, 40, 225–254. [Google Scholar]
- Nicholson, G.M. Insect-selective spider toxins targeting voltage-gated sodium channels. Toxicon 2007, 49, 490–512. [Google Scholar]
- De Lima, M.E.; Figueiredo, S.G.; Pimenta, A.M.; Santos, D.M.; Borges, M.H.; Cordeiro, M.N.; Richardson, M.; Oliveira, L.C.; Stankiewicz, M.; Pelhate, M. Peptides of arachnid venoms with insecticidal activity targeting sodium channels. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2007, 146, 264–279. [Google Scholar]
- Sollod, B.L.; Wilson, D.; Zhaxybayeva, O.; Gogarten, J.P.; Drinkwater, R.; King, G.F. Were arachnids the first to use combinatorial peptide libraries? Peptides 2005, 26, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Grishin, E. Polypeptide neurotoxins from spider venoms. Eur. J. Biochem. 1999, 264, 276–280. [Google Scholar]
- Escoubas, P.; Diochot, S.; Corzo, G. Structure and pharmacology of spider venom neurotoxins. Biochimie 2000, 82, 893–907. [Google Scholar]
- Dutra, A.A.; Sousa, L.O.; Resende, R.R.; Brandao, R.L.; Kalapothakis, E.; Castro, I.M. Expression and characterization of LTx2, a neurotoxin from Lasiodora sp. effecting on calcium channels. Peptides 2008, 29, 1505–1513. [Google Scholar] [CrossRef] [PubMed]
- Norton, R.S.; Pallaghy, P.K. The cystine knot structure of ion channel toxins and related polypeptides. Toxicon 1998, 36, 1573–1583. [Google Scholar]
- Schalle, J.; Kampfer, U.; Schurch, S.; Kuhn-Nentwig, L.; Haeberli, S.; Nentwig, W. CSTX-9, a toxic peptide from the spider Cupiennius salei: amino acid sequence, disulphide bridge pattern and comparison with other spider toxins containing the cystine knot structure. Cell Mol. Life Sci. 2001, 58, 1538–1545. [Google Scholar] [CrossRef] [PubMed]
- Mouhat, S.; Jouirou, B.; Mosbah, A.; De Waard, M.; Sabatier, J.M. Diversity of folds in animal toxins acting on ion channels. Biochem. J. 2004, 378, 717–726. [Google Scholar]
- Corzo, G.; Escoubas, P. Pharmacologically active spider peptide toxins. Cell Mol. Life Sci. 2003, 60, 2409–2426. [Google Scholar]
- Corzo, G.; Escoubas, P.; Stankiewicz, M.; Pelhate, M.; Kristensen, C.P.; Nakajima, T. Isolation, synthesis and pharmacological characterization of δ-palutoxins IT, novel insecticidal toxins from the spider Paracoelotes luctuosus (Amaurobiidae). Eur. J. Biochem. 2000, 267, 5783–5795. [Google Scholar] [CrossRef] [PubMed]
- Tedford, H.W.; Sollod, B.L.; Maggio, F.; King, G.F. Australian funnel-web spiders: master insecticide chemists. Toxicon 2004, 43, 601–618. [Google Scholar]
- Black, B.C.; Brennam, L.A.; Dierks, P.M.; Gard, I.E. Commercialization of baculoviral insecticides. In The Baculoviruses (Miller,Lois). In The Viruses; Plenum Press: New York, NY, USA, 1997; pp. 341–347. [Google Scholar]
- Neurath, H. Proteolytic processing and physiological regulation. Trends Biochem. Sci. 1989, 14, 268–271. [Google Scholar]
- Otlewski, J.; Krowarsch, D.; Apostoluk, W. Protein inhibitors of serine proteinases. Acta Biochim. Pol. 1999, 46, 531–565. [Google Scholar]
- Rimphanitchayakit, V.; Tassanakajon, A. Structure and function of invertebrate Kazal-type serine proteinase inhibitors. Dev. Comp. Immunol. 2010, 34, 377–386. [Google Scholar]
- Laskowski, M., Jr.; Kato, I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980, 49, 593–626. [Google Scholar]
- Krowarsch, D.; Cierpicki, T.; Jelen, F.; Otlewski, J. Canonical protein inhibitors of serine proteases. Cell Mol. Life Sci. 2003, 60, 2427–2444. [Google Scholar]
- Irving, J.A.; Pike, R.N.; Lesk, A.M.; Whisstock, J.C. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Res. 2000, 10, 1845–1864. [Google Scholar]
- Law, R.H.; Zhang, Q.; McGowan, S.; Buckle, A.M.; Silverman, G.A.; Wong, W.; Rosado, C.J.; Langendorf, C.G.; Pike, R.N.; Bird, P.I.; Whisstock, J.C. An overview of the serpin superfamily. Genome Biol. 2006, 7, 216. [Google Scholar]
- Takahashi, H.; Iwanaga, S.; Suzuki, T. Distribution of proteinase inhibitors in snake venoms. Toxicon 1974, 12, 193–197. [Google Scholar]
- Shafqat, J.; Beg, O.U.; Yin, S.J.; Zaidi, Z.H.; Jornvall, H. Primary structure and functional properties of cobra (Naja naja naja) venom Kunitz-type trypsin inhibitor. Eur. J. Biochem. 1990, 194, 337–341. [Google Scholar]
- Shafqat, J.; Zaidi, Z.H.; Jornvall, H. Purification and characterization of a chymotrypsin Kunitz inhibitor type of polypeptide from the venom of cobra (Naja naja naja). FEBS Lett. 1990, 275, 6–8. [Google Scholar]
- Chang, L.; Chung, C.; Huang, H.B.; Lin, S. Purification and characterization of a chymotrypsin inhibitor from the venom of Ophiophagus hannah (King Cobra). Biochem. Biophys. Res. Commun. 2001, 283, 862–867. [Google Scholar]
- Chen, C.; Hsu, C.H.; Su, N.Y.; Lin, Y.C.; Chiou, S.H.; Wu, S.H. Solution structure of a Kunitz-type chymotrypsin inhibitor isolated from the elapid snake Bungarus fasciatus. J. Biol. Chem. 2001, 276, 45079–45087. [Google Scholar]
- Lu, X.Z.; Zou, Y.G.; Yin, X.M.; Chen, W.T.; Zhang, C.P. Expression of MMP1 mRNA in oral squamous cell carcinoma and paired normal tissues. Nan Fang Yi Ke Da Xue Xue Bao 2008, 28, 1362–1364. [Google Scholar]
- Zhou, X.D.; Jin, Y.; Lu, Q.M.; Li, D.S.; Zhu, S.W.; Wang, W.Y.; Xiong, Y.L. Purification, characterization and primary structure of a chymotrypsin inhibitor from Naja atra venom. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2004, 137, 219–224. [Google Scholar]
- He, D.; Natarajan, V.; Stern, R.; Gorshkova, I.A.; Solway, J.; Spannhake, E.W.; Zhao, Y. Lysophosphatidic acid-induced transactivation of epidermal growth factor receptor regulates cyclo-oxygenase-2 expression and prostaglandin E(2) release via C/EBPbeta in human bronchial epithelial cells. Biochem. J. 2008, 412, 153–162. [Google Scholar]
- Millers, E.K.; Trabi, M.; Masci, P.P.; Lavin, M.F.; de Jersey, J.; Guddat, L.W. Crystal structure of textilinin-1, a Kunitz-type serine protease inhibitor from the venom of the Australian common brown snake (Pseudonaja textilis). FEBS J. 2009, 276, 3163–3175. [Google Scholar] [CrossRef]
- Flight, S.M.; Johnson, L.A.; Trabi, M.; Gaffney, P.; Lavin, M.; de Jersey, J.; Masci, P. Comparison of textilinin-1 with aprotinin as serine protease inhibitors and as antifibrinolytic agents. Pathophysiol. Haemost. Thromb. 2005, 34, 188–193. [Google Scholar]
- Flight, S.M.; Johnson, L.A.; Du, Q.S.; Warner, R.L.; Trabi, M.; Gaffney, P.J.; Lavin, M.F.; de Jersey, J.; Masci, P.P. Textilinin-1, an alternative anti-bleeding agent to aprotinin: Importance of plasmin inhibition in controlling blood loss. Br. J. Haematol. 2009, 145, 207–211. [Google Scholar]
- Zhao, Y.; Jin, Y.; Wei, S.S.; Lee, W.H.; Zhang, Y. Purification and characterization of an irreversible serine protease inhibitor from skin secretions of Bufo andrewsi. Toxicon 2005, 46, 635–640. [Google Scholar]
- Yuan, C.H.; He, Q.Y.; Peng, K.; Diao, J.B.; Jiang, L.P.; Tang, X.; Liang, S.P. Discovery of a distinct superfamily of Kunitz-type toxin (KTT) from tarantulas. PLoS One 2008, 3, e3414. [Google Scholar]
- Duarte, M.M.; Montes De Oca, H.; Diniz, C.R.; Fortes-Dias, C.L. Primary culture of venom gland cells from the South American rattlesnake (Crotalus durissus terrificus). Toxicon 1999, 37, 1673–1682. [Google Scholar]
- Yamanouye, N.; Kerchove, C.M.; Moura-da-Silva, A.M.; Carneiro, S.M.; Markus, R.P. Long-term primary culture of secretory cells of Bothrops jararaca venom gland for venom production in vitro. Nat. Protocols 2007, 1, 2763–2766. [Google Scholar]
- Silva, L.M.; Lages, C.P.; Venuto, T.; Lima, R.M.; Diniz, M.V.; Valentim, C.L.L.; Baba, E.H.; Pimenta, P.F.P.; Fortes-Dias, C.L. Primary culture of venom glands from the Brazilian armed spider, Phoneutria nigriventer (Araneae, Ctenidae). Toxicon 2008, 51, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Domingos, M.O.; Barbaro, K.C.; Tynan, W.; Penny, J.; Lewis, D.J.; New, R.R. Influence of sphingomyelin and TNF-alpha release on lethality and local inflammatory reaction induced by Loxosceles gaucho spider venom in mice. Toxicon 2003, 42, 471–479. [Google Scholar]
- Barbaro, K.C.; Lira, M.S.; Araujo, C.A.; Pareja-Santos, A.; Tavora, B.C.; Prezotto-Neto, J.P.; Kimura, L.F.; Lima, C.; Lopes-Ferreira, M.; Santoro, M.L. Inflammatory mediators generated at the site of inoculation of Loxosceles gaucho spider venom. Toxicon 2010, 56, 972–979. [Google Scholar]
- Burgess, R.R.; Richard, R.B.; Murray, P.D. Refolding Solubilized Inclusion Body Proteins. In Methods in Enzymology; Academic Press: Salt Lake City, UT, USA, 2009; Volume 463, pp. 259–282. Chapter. 17. [Google Scholar]
- Daly, R.; Hearn, M.T. Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J. Mol. Recognit. 2005, 18, 119–138. [Google Scholar]
- Benting, J.; Lecat, S.; Zacchetti, D.; Simons, K. Protein Expression in Drosophila Schneider Cells. Anal. Biochem. 2000, 278, 59–68. [Google Scholar]
- Rohrmann, G.F. Baculovirus Molecular Biology; European Molecular Biology Organization: Corvallis, OR, USA, 2008. [Google Scholar]
- Wurm, F.M. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat. Biotech. 2004, 22, 1393–1398. [Google Scholar]
- Escoubas, P.; Sollod, B.; King, G.F. Venom landscapes: Mining the complexity of spider venoms via a combined cDNA and mass spectrometric approach. Toxicon 2006, 47, 650–663. [Google Scholar]
- Murakami, M.T.; Fernandes-Pedrosa, M.F.; Tambourgi, D. V.; Arni, R.K. Structural basis for metal ion coordination and the catalytic mechanism of sphingomyelinases D. J. Biol. Chem. 2005, 280, 13658–13664. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chaim, O.M.; Trevisan-Silva, D.; Chaves-Moreira, D.; Wille, A.C.M.; Ferrer, V.P.; Matsubara, F.H.; Mangili, O.C.; Silveira, R.B.d.; Gremski, L.H.; Gremski, W.; et al. Brown Spider (Loxosceles genus) Venom Toxins: Tools for Biological Purposes. Toxins 2011, 3, 309-344. https://doi.org/10.3390/toxins3030309
Chaim OM, Trevisan-Silva D, Chaves-Moreira D, Wille ACM, Ferrer VP, Matsubara FH, Mangili OC, Silveira RBd, Gremski LH, Gremski W, et al. Brown Spider (Loxosceles genus) Venom Toxins: Tools for Biological Purposes. Toxins. 2011; 3(3):309-344. https://doi.org/10.3390/toxins3030309
Chicago/Turabian StyleChaim, Olga Meiri, Dilza Trevisan-Silva, Daniele Chaves-Moreira, Ana Carolina M. Wille, Valéria Pereira Ferrer, Fernando Hitomi Matsubara, Oldemir Carlos Mangili, Rafael Bertoni da Silveira, Luiza Helena Gremski, Waldemiro Gremski, and et al. 2011. "Brown Spider (Loxosceles genus) Venom Toxins: Tools for Biological Purposes" Toxins 3, no. 3: 309-344. https://doi.org/10.3390/toxins3030309
APA StyleChaim, O. M., Trevisan-Silva, D., Chaves-Moreira, D., Wille, A. C. M., Ferrer, V. P., Matsubara, F. H., Mangili, O. C., Silveira, R. B. d., Gremski, L. H., Gremski, W., Senff-Ribeiro, A., & Veiga, S. S. (2011). Brown Spider (Loxosceles genus) Venom Toxins: Tools for Biological Purposes. Toxins, 3(3), 309-344. https://doi.org/10.3390/toxins3030309