Gene Expression Profiling and Identification of Resistance Genes to Aspergillus flavus Infection in Peanut through EST and Microarray Strategies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Peanut Lines Used
2.2. Peanut Inoculation by Aspergillus during Growth
2.3. Expressed Sequence Tags and Sequencing
2.4. Oligo Microarray Design
2.5. Microarray Experiment Design, Hybridization and Analysis
- C20Y vs. TFY (GT-C20 infected vs. Tifrunner infected)
- C20Y vs. C20N (GT-C20 infected vs. not infected)
- TFY vs. TFN (Tifrunner infected vs. not infected)
- C20N vs. TFN (GT-C20 not infected vs. Tifrunner not infected)
2.6. Data Processing for EST and Microarray Analysis
3. Results and Discussion
3.1. Summary Classification of Expressed Sequence Tags(EST)
| Category of Genes | Number of Genes |
|---|---|
| Hypothetical proteins | 12,118 |
| Ribosomal protein | 131 |
| Lopprotein | 91 |
| Cupin | 54 |
| Ribulose bisphophate carboxylase | 36 |
| Oleisin | 33 |
| Conglutin | 32 |
| Photosystem I and II | 29 |
| Protease inhibitor/seed storage protein | 28 |
| Core histone | 25 |
| Ara H8 allergen/alergen | 25 |
| Ubiquitin-conjugating enzyme | 23 |
| Peptidases | 22 |
| Epoxide hydrolase | 19 |
| Ras family protein | 16 |
| Glutathionine S-transferase | 16 |
| Zinc figure protein | 14 |
| Seed maturation protein | 13 |
| NAD/NADH dehydrogenase | 12 |
| Mem brane protein | 12 |
| Hsp20 | 11 |
| Peroxidase | 10 |
| 14-3-3 protein | 10 |
| Universal stress protein | 9 |
| Oxidoreductase | 9 |
| HMG(high mobility group) box | 8 |
| Protein kinase | 6 |
| Polygalacturonase | 4 |
| Other | 1063 |
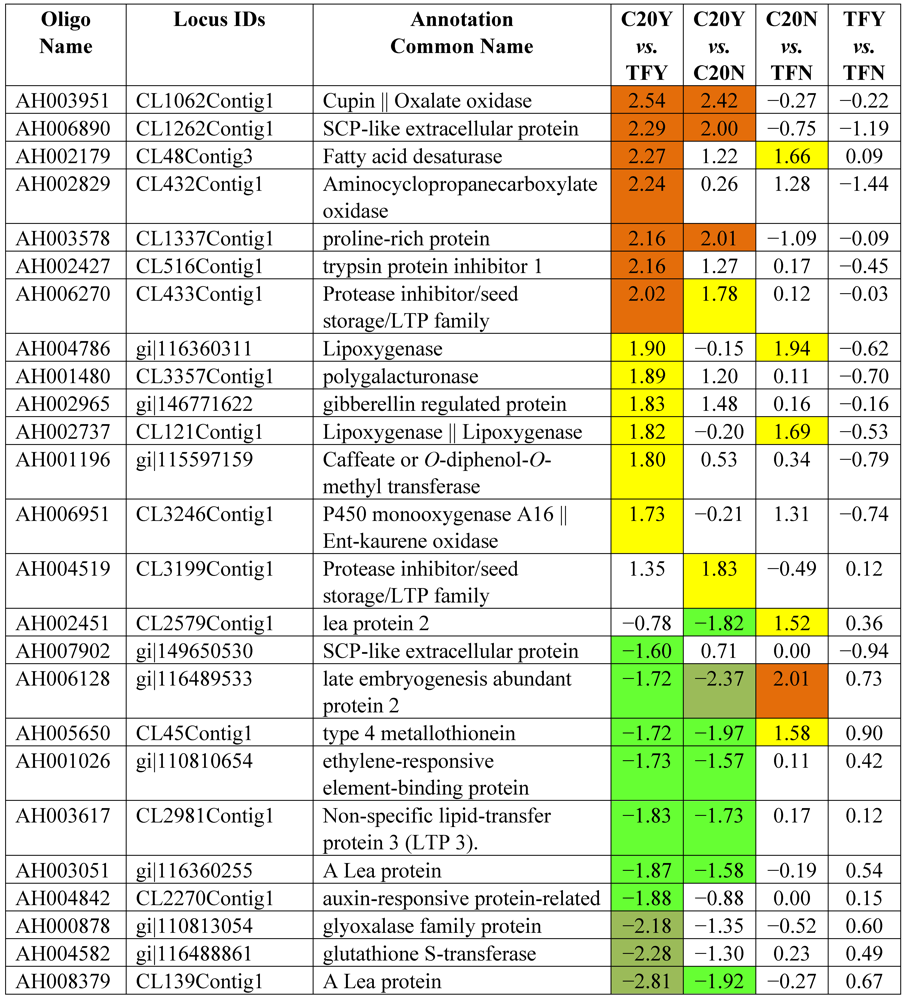
3.2. Identification of Resistant Genes to Aspergillus Infection Using Microarray Expression Data
| Differential Expression | ||||
|---|---|---|---|---|
| Hybridizations | Up-high (Log2 ≥ 2) | Up-mod (Log2 ≥ 1.5 &< 2) | Down-high (Log2 ≤ −2) | Down-mod (Log2 ≤ −1.5 & > −2) |
| C20Y vs. TFY | 52 | 126 | 51 | 99 |
| C20Y vs. C20N | 25 | 40 | 9 | 38 |
| C20N vs. TFN | 9 | 31 | 3 | 19 |
| TFY vs. TFN | 0 | 1 | 0 | 4 |
 |
 |
 |
 |
 |
3.3. Genes Resistant to Fungal Infection in Other Crop Systems have been Identified
3.4. Defense-Related Genes Identified by Peanut Seed EST Database Search
4. Conclusions
Acknowledgements
References
- Guo, B.Z.; Chen, C.Y.; Chu, Y.; Holbrook, C.C.; Ozias-Akins, P.; Stalker, H.T. Advances in Genetics and Genomics for Sustainable Peanut Production. In Sustainable Agriculture and New Biotechnologies; Benkeblia, N., Ed.; CRC Press: Boca Raton, FL, USA, 2011; pp. 341–368. [Google Scholar]
- Guo, B.Z.; Yu, J.; Holbrook, C.C.; Cleveland, T.E.; Nierman, W.C.; Scully, B.T. Strategy in prevention of prehavest aflatoxin contamination in peanuts: Aflatoxin biosynthesis, genetics and genomics. Peanut Sci. 2009, 36, 11–20. [Google Scholar]
- Peanut Biosciences. Available online: http://www.peanutbioscience.com/peanutgenomeinitiative.html (accessed on 23 June 2011).
- Guo, B.; Chen, X.; Hong, Y.; Liang, X.; Dang, P.; Brenneman, T.; Holbrook, C.; Culbreath, A. Analysis of gene expression profiles in leaf tissues of cultivated peanuts and development of EST-SSR markers and gene discovery. Int. J. Plant Genomics 2009, 715605:1–715605:14. [Google Scholar]
- Guo, B.; Chen, X.; Dang, P.; Scully, B.T.; Liang, X.; Holbrook, C.C.; Yu, J.; Culbreath, A.K. Peanut gene expression profiling in developing seeds at different reproduction stages during Aspergillus parasiticus infection. BMC Dev. Biol. 2008, 8, 12. [Google Scholar]
- Luo, M.; Dang, P.; Guo, B.Z.; He, G.; Holbrook, C.C.; Bausher, M.G.; Lee, R.D. Generation of expressed sequence tags (ESTs) for gene discovery and marker development in cultivated peanut. Crop Sci. 2005, 45, 346–353. [Google Scholar]
- Luo, M.; Dang, P.; Bausher, M.G.; Holbrook, C.C.; Lee, R.D.; Lynch, R.E.; Guo, B.Z. Identification of transcripts involved in resistance responses to leaf spot disease caused by cercosporidium personatum in peanut (Arachis hypogaea). Phytopathol. 2005, 95, 381–387. [Google Scholar]
- Luo, M.; Liang, X.Q.; Dang, P.; Holbrook, C.C.; Bausher, M.G.; Lee, R.D.; Guo, B.Z. Microarray-based screening of differentially expressed genes in peanut in response to Aspergillus parasiticus infection and drought stress. Plant Sci. 2005, 169, 695–703. [Google Scholar]
- Haegeman, A.; Jacob, J.; Vanholme, B.; Kyndt, T.; Mitreva, M.; Gheysen, G. Expressed sequence tags of the peanut pod nematode Ditylenchus africanus: the first transcriptome analysis of an Anguinid nematode. Mol. Biochem. Parasitol. 2009, 167, 32–40. [Google Scholar]
- Tirumalaraju, S.V.; Jain, M.; Gallo, M. Differential gene expression in roots of nematode-resistant and -susceptible peanut (Arachis hypogaea) cultivars in response to early stages of peanut root-knot nematode (Meloidogyne arenaria) parasitization. J. Plant Physiol. 2011, 168, 481–492. [Google Scholar]
- Bi, Y.P.; Liu, W.; Xia, H.; Su, L.; Zhao, C.Z.; Wan, S.B.; Wang, X.J. EST sequencing and gene expression profiling of cultivated peanut (Arachis hypogaea L.). Genome 2010, 53, 832–839. [Google Scholar] [CrossRef] [PubMed]
- Payton, P.; Kottapalli, K.R.; Rowland, D.; Faircloth, W.; Guo, B.; Burow, M.; Puppala, N.; Gallo, M. Gene expression profiling in peanut using high density oligonucleotide microarrays. BMC Genomics 2009, 10, 265. [Google Scholar]
- Guo, B.Z.; Chen, X.P.; Dang, P.; Scully, B.T.; Liang, X.Q.; Holbrook, C.C.; Yu, J.; Culbreath, A.K. Peanut gene expression profiling in developing seeds at different reproduction stages during Aspergillus parasiticus infection. BMC Dev. Biol. 2008, 8, 12. [Google Scholar]
- National Center for Biotechnology Information. Available online: http://www.ncbi.nlm.nih.gov (accesed on 23 June 2011).
- Holbrook, C.C.; Culbreath, A.K. Registration of “Tifrunner” Peanut. J. Plant Regist. 2007, 1, 124. [Google Scholar]
- Liang, X.Q.; Holbrook, C.C.; Lynch, R.E.; Guo, B.Z. Beta-1,3-glucanase activity in peanut seed (Arachis hypogaea) is induced by inoculation with Aspergillus flavus and copurifies with a conglutin-like protein. Phytopathology 2005, 95, 506–511. [Google Scholar]
- Mixon, A.C. Reducing Aspergillus species infection of peanut seed using resistant genotypes. J. Environ. Qual. 1986, 15, 101–103. [Google Scholar]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar]
- Paracel BLAST. Available online: http://www.paracel.com (accessed on 23 June 2011).
- Wilkinson, J.R.; Yu, J.; Abbas, H.K.; Scheffler, B.E.; Kim, H.S.; Nierman, W.C.; Bhatnagar, D.; Cleveland, T.E. Aflatoxin formation and gene expression in response to carbon source media shift in Aspergillus parasiticus. J. Food Addit. Contam. 2007, 24, 1051–1060. [Google Scholar]
- Wilkinson, J.R.; Yu, J.; Bland, J.M.; Nierman, W.C.; Bhatnagar, D.; Cleveland, T.E. Amino acid supplementation reveals differential regulation of aflatoxin biosynthesis in Aspergillus flavus NRRL 3357 and Aspergillus parasiticus SRRC 143. Appl. Microbiol. Biotechnol. 2007, 74, 1308–1319. [Google Scholar]
- Multi Experiment Viewer. Available online: http://www.tm4.org/mev.html (accessed on 23 June 2011).
- J Craig Venter Institute Metagenomics Reports (METAREP). Available online: http://www.jcvi.org/metarep/dashboard/index (accessed on 23 June 2011).
- Banks, W.A.; Niehoff, M.L.; Brown, R.L.; Chen, Z.Y.; Cleveland, T.E. Transport of an antifungal trypsin inhibitor isolated from corn across the blood-brain barrier. Antimicrob. Agents Chemother. 2002, 46, 2633–2635. [Google Scholar]
- Chen, Z.Y.; Brown, R.L.; Lax, A.R.; Cleveland, T.E.; Russin, J.S. Inhibition of plant-pathogenic fungi by a corn trypsin inhibitor overexpressed in Escherichia coli. Appl. Environ. Microbiol. 1999, 65, 1320–1324. [Google Scholar]
- Chen, Z.Y.; Brown, R.L.; Russin, J.S.; Lax, A.R.; Cleveland, T.E. A corn trypsin inhibitor with antifungal activity inhibits aspergillus flavus alpha-amylase. Phytopathology 1999, 89, 902–907. [Google Scholar]
- Guo, B.Z.; Brown, R.L.; Lax, A.R.; Cleveland, T.E.; Russin, J.S.; Widstrom, N.W. Protein profiles and antifungal activities of kernel extracts from corn genotypes resistant and susceptible to Aspergillus flavus. J. Food Prot. 1998, 61, 98–102. [Google Scholar]
- Burow, G.B.; Gardner, H.W.; Keller, N.P. A peanut seed lipoxygenase responsive to Aspergillus colonization. Plant Mol. Biol. 2000, 42, 689–701. [Google Scholar]
- Burow, G.B.; Nesbitt, J.D.; Keller, N.P. Seed lipoxygenase products modulate Aspergillus mycotoxin synthesis. Mol. Plant Microbe Interact. 1997, 10, 380–387. [Google Scholar]
- Wilson, R.A.; Gardner, H.W.; Keller, N.P. Cultivar-dependent expression of a maize lipoxygenase responsive to seed infesting fungi. Mol. Plant Microbe Interact. 2001, 14, 980–987. [Google Scholar]
- Calvo, A.M.; Hinze, L.L.; Gardner, H.W.; Keller, N.P. Sporogenic effect of polyunsaturated fatty acids on development of Aspergillus spp. Appl. Environ. Microbiol. 1999, 65, 3668–3673. [Google Scholar]
- Gao, X.; Brodhagen, M.; Isakeit, T.; Brown, S.H.; Gobel, C.; Betran, J.; Feussner, I.; Keller, N.P.; Kolomiets, M.V. Inactivation of the lipoxygenase ZmLOX3 increases susceptibility of maize to Aspergillus spp. Mol. Plant Microbe Interact. 2009, 22, 222–231. [Google Scholar]
- Roy, S.K.; Kulkarni, A.P. Aflatoxin B1 epoxidation catalysed by partially purified human liver lipoxygenase. Xenobiotica 1997, 27, 231–241. [Google Scholar]
- Datta, K.; Kulkarni, A.P. Oxidative metabolism of aflatoxin B1 by lipoxygenase purified from human term placenta and intrauterine conceptal tissues. Teratology 1994, 50, 311–317. [Google Scholar]
- Stekel, D.J.; Git, Y.; Falciani, F. The comparison of gene expression from multiple cDNA libraries. Genome Res. 2000, 10, 2055–2061. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Guo, B.; Fedorova, N.D.; Chen, X.; Wan, C.-H.; Wang, W.; Nierman, W.C.; Bhatnagar, D.; Yu, J. Gene Expression Profiling and Identification of Resistance Genes to Aspergillus flavus Infection in Peanut through EST and Microarray Strategies. Toxins 2011, 3, 737-753. https://doi.org/10.3390/toxins3070737
Guo B, Fedorova ND, Chen X, Wan C-H, Wang W, Nierman WC, Bhatnagar D, Yu J. Gene Expression Profiling and Identification of Resistance Genes to Aspergillus flavus Infection in Peanut through EST and Microarray Strategies. Toxins. 2011; 3(7):737-753. https://doi.org/10.3390/toxins3070737
Chicago/Turabian StyleGuo, Baozhu, Natalie D. Fedorova, Xiaoping Chen, Chun-Hua Wan, Wei Wang, William C. Nierman, Deepak Bhatnagar, and Jiujiang Yu. 2011. "Gene Expression Profiling and Identification of Resistance Genes to Aspergillus flavus Infection in Peanut through EST and Microarray Strategies" Toxins 3, no. 7: 737-753. https://doi.org/10.3390/toxins3070737
APA StyleGuo, B., Fedorova, N. D., Chen, X., Wan, C.-H., Wang, W., Nierman, W. C., Bhatnagar, D., & Yu, J. (2011). Gene Expression Profiling and Identification of Resistance Genes to Aspergillus flavus Infection in Peanut through EST and Microarray Strategies. Toxins, 3(7), 737-753. https://doi.org/10.3390/toxins3070737





