Adapting Yeast as Model to Study Ricin Toxin A Uptake and Trafficking †
Abstract
:Abbreviations
| ER | endoplasmic reticulum |
| RTA | ricin toxin A chain |
| RTB | ricin toxin B chain |
| PI | propidiumiodide |
1. Introduction
2. Materials and Methods
2.1. Escherichia Coli Strains, Plasmids, Culture Media and Genetic Techniques
2.2. Yeast Strains and Culture Media
2.3. Affinity Purification of RTA
2.4. Western Analysis and Protein Staining
2.5. Phase Contrast Microscopy
2.6. Oyxgen-Sensor Microtiter Plate Bioassay


2.7. Cytotoxicity Assay
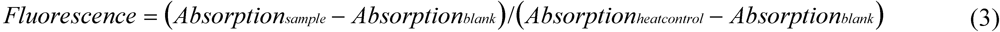
2.8. Cell Culture

3. Results and Discussion
3.1. RTA Expression and Purification

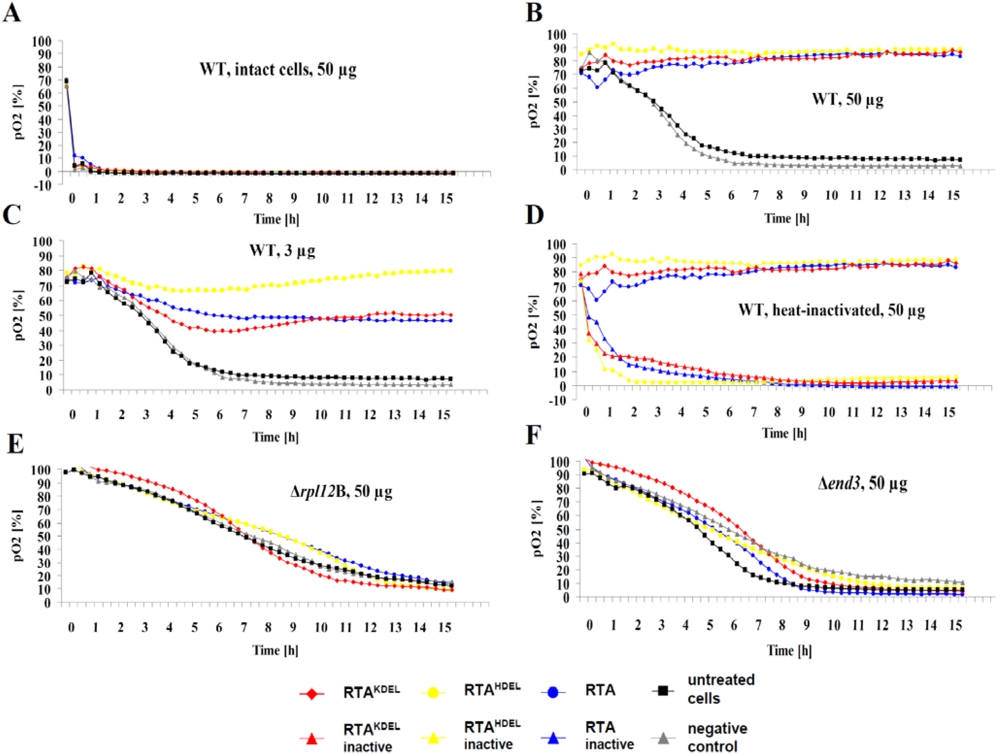
3.2. RTA Containing a Mammalian or Yeast Specific ER Retention Signal is in vivo Toxic

3.3. Recombinant RTA Variants Induce Cell Death in Yeast Spheroplasts
3.4. RTA Endocytosis is an Essential Prerequisite for Cell Killing

4. Conclusions
Acknowlegments
References
- Montanaro, L.; Sperti, S.; Stirpe, F. Inhibition by ricin of protein synthesis in vitro. Ribosomes as the target of the toxin. Biochem. J. 1973, 136, 677–683. [Google Scholar] [PubMed]
- Day, P.J.; Owens, S.R.; Wesche, J.; Olsnes, S.; Roberts, L.M.; Lord, J.M. An interaction between ricin and calreticulin that may have implications for toxin trafficking. J. Biol. Chem. 2001, 276, 7202–7208. [Google Scholar]
- Weston, S.A.; Tucker, A.D.; Thatcher, D.R.; Derbyshire, D.J.; Pauptit, R.A. X-ray structure of recombinant ricin A-chain at 1.8 A resolution. J. Mol. Biol. 1994, 244, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Tsurugi, K. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J. Biol. Chem. 1987, 262, 8128–8130. [Google Scholar]
- Szewczak, A.A.; Moore, P.B.; Chang, Y.L.; Wool, I.G. The conformation of the sarcin/ricin loop from 28S ribosomal RNA. Proc. Natl. Acad. Sci. USA 1993, 90, 9581–9585. [Google Scholar]
- Moazed, D.; Robertson, J.M.; Noller, H.F. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature 1988, 334, 362–364. [Google Scholar]
- Newton, D.L.; Wales, R.; Richardson, P.T.; Walbridge, S.; Saxena, S.K.; Ackerman, E.J.; Roberts, L.M.; Lord, J.M.; Youle, R.J. Cell surface and intracellular functions for ricin galactose binding. J. Biol. Chem. 1992, 267, 11917–11922. [Google Scholar]
- Sandvig, K.; Olsnes, S. Entry of the toxic proteins abrin, modeccin, ricin, and diphtheria toxin into cells. II. Effect of pH, metabolic inhibitors, and ionophores and evidence for toxin penetration from endocytotic vesicles. J. Biol. Chem. 1982, 257, 7504–7513. [Google Scholar] [PubMed]
- Moya, M.; Dautry-Varsat, A.; Goud, B.; Louvard, D.; Boquet, P. Inhibition of coated pit formation in Hep2 cells blocks the cytotoxicity of diphtheria toxin but not that of ricin toxin. J. Cell Biol. 1985, 101, 548–559. [Google Scholar]
- Iversen, T.G.; Skretting, G.; Llorente, A.; Nicoziani, P.; van Deurs, B.; Sandvig, K. Endosome to Golgi transport of ricin is independent of clathrin and of the Rab9- and Rab11-GTPases. Mol. Biol. Cell 2001, 12, 2099–2107. [Google Scholar]
- Sandvig, K.; van Deurs, B. Endocytosis, intracellular transport, and cytotoxic action of Shiga toxin and ricin. Physiol. Rev. 1996, 76, 949–966. [Google Scholar]
- van Deurs, B.; Sandvig, K.; Petersen, O.W.; Olsnes, S.; Simons, K.; Griffiths, G. Estimation of the amount of internalized ricin that reaches the trans-Golgi network. J. Cell Biol. 1988, 106, 253–267. [Google Scholar]
- Stechmann, B.; Bai, S.K.; Gobbo, E.; Lopez, R.; Merer, G.; Pinchard, S.; Panigai, L.; Tenza, D.; Raposo, G.; Beaumelle, B.; et al. Inhibition of retrograde transport protects mice from lethal ricin challenge. Cell 2010, 141, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Lord, J.M.; Roberts, L.M.; Lencer, W.I. Entry of protein toxins into mammalian cells by crossing the endoplasmic reticulum membrane: Co-opting basic mechanisms of endoplasmic reticulum-associated degradation. Curr. Top. Microbiol. Immunol. 2005, 300, 149–168. [Google Scholar]
- Lord, J.M.; Roberts, L.M.; Robertus, J.D. Ricin: Structure, mode of action, and some current applications. FASEB J. 1994, 8, 201–208. [Google Scholar] [PubMed]
- Simpson, J.C.; Roberts, L.M.; Romisch, K.; Davey, J.; Wolf, D.H.; Lord, J.M. Ricin A chain utilises the endoplasmic reticulum-associated protein degradation pathway to enter the cytosol of yeast. FEBS Lett. 1999, 459, 80–84. [Google Scholar]
- Sokolowska, I.; Walchli, S.; Wegrzyn, G.; Sandvig, K.; Slominska-Wojewodzka, M. A single point mutation in ricin A-chain increases toxin degradation and inhibits EDEM1-dependent ER retrotranslocation. Biochem. J. 2011, 2, 371–385. [Google Scholar]
- Slominska-Wojewodzka, M.; Gregers, T.F.; Walchli, S.; Sandvig, K. EDEM is involved in retrotranslocation of ricin from the endoplasmic reticulum to the cytosol. Mol. Biol. Cell 2006, 17, 1664–1675. [Google Scholar]
- Lord, M.J.; Jolliffe, N.A.; Marsden, C.J.; Pateman, C.S.; Smith, D.C.; Spooner, R.A.; Watson, P.D.; Roberts, L.M. Ricin. Mechanisms of cytotoxicity. Toxicol. Rev. 2003, 22, 53–64. [Google Scholar] [PubMed]
- Zhou, X.X.; Ji, F.; Zhao, J.L.; Cheng, L.F.; Xu, C.F. Anti-cancer activity of anti-p185HER-2 ricin A chain immunotoxin on gastric cancer cells. J. Gastroenterol. Hepatol. 2010, 25, 1266–1275. [Google Scholar]
- Surash, S.; Nemeth, P.; Chakrabarty, A.; Chumas, P. The conjugation of an AQP1-directed immunotoxin in the study of site-directed therapy within the CNS. Childs Nerv. Syst. 2011, 27, 811–818. [Google Scholar]
- Mathew, M.; Verma, R.S. Humanized immunotoxins: A new generation of immunotoxins for targeted cancer therapy. Cancer Sci. 2009, 100, 1359–1365. [Google Scholar]
- Bradley, J.L.; Silva, H.M.; McGuire, P.M. Depurination of yeast 26S ribosomal RNA by recombinant ricin A chain. Biochem. Biophys. Res. Commun. 1987, 149, 588–593. [Google Scholar]
- Li, S.; Spooner, R.A.; Allen, S.C.; Guise, C.P.; Ladds, G.; Schnoder, T.; Schmitt, M.J.; Lord, J.M.; Roberts, L.M. Folding-competent and folding-defective forms of ricin A chain have different fates after retrotranslocation from the endoplasmic reticulum. Mol. Biol. Cell 2010, 21, 2543–2554. [Google Scholar]
- Wales, R.; Roberts, L.M.; Lord, J.M. Addition of an endoplasmic reticulum retrieval sequence to ricin A chain significantly increases its cytotoxicity to mammalian cells. J. Biol. Chem. 1993, 268, 23986–23990. [Google Scholar]
- Wales, R.; Chaddock, J.A.; Roberts, L.M.; Lord, J.M. Addition of an ER retention signal to the ricin A chain increases the cytotoxicity of the holotoxin. Exp. Cell Res. 1992, 203, 1–4. [Google Scholar]
- Sambrook, J.; Maniatis, T.; Fritsch, E.F. Molecular Cloning: A Laboratory Manual, 2nd ed; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Schagger, H.; von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar]
- Schagger, H.; Aquila, H.; von Jagow, G. Coomassie blue-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for direct visualization of polypeptides during electrophoresis. Anal. Biochem. 1988, 173, 201–205. [Google Scholar]
- John, G.T.; Klimant, I.; Wittmann, C.; Heinzle, E. Integrated optical sensing of dissolved oxygen in microtiter plates: A novel tool for microbial cultivation. Biotechnol. Bioeng. 2003, 81, 829–836. [Google Scholar]
- Allen, S.C.; Moore, K.A.; Marsden, C.J.; Fulop, V.; Moffat, K.G.; Lord, J.M.; Ladds, G.; Roberts, L.M. The isolation and characterization of temperature-dependent ricin A chain molecules in Saccharomyces cerevisiae. FEBS J. 2007, 274, 5586–5599. [Google Scholar]
- Zhan, J.; de Sousa, M.; Chaddock, J.A.; Roberts, L.M.; Lord, J.M. Restoration of lectin activity to a non-glycosylated ricin B chain mutant by the introduction of a novel N-glycosylation site. FEBS Lett. 1997, 407, 271–274. [Google Scholar] [CrossRef]
- Wang, M.; Yu, S.; Wang, C.; Kong, J. Tracking the endocytic pathway of recombinant protein toxin delivered by multiwalled carbon nanotubes. ACS Nano 2010, 4, 6483–6490. [Google Scholar]
- Raykhel, I.; Alanen, H.; Salo, K.; Jurvansuu, J.; Nguyen, V.D.; Latva-Ranta, M.; Ruddock, L. A molecular specificity code for the three mammalian KDEL receptors. J. Cell Biol. 2007, 179, 1193–1204. [Google Scholar]
- Semenza, J.C.; Hardwick, K.G.; Dean, N.; Pelham, H.R. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell 1990, 61, 1349–1357. [Google Scholar]
- Semenza, J.C.; Pelham, H.R. Changing the specificity of the sorting receptor for luminal endoplasmic reticulum proteins. J. Mol. Biol. 1992, 224, 1–5. [Google Scholar]
- Eiklid, K.; Olsnes, S.; Pihl, A. Entry of lethal doses of abrin, ricin and modeccin into the cytosol of HeLa cells. Exp. Cell Res. 1980, 126, 321–326. [Google Scholar]
- Raths, S.; Rohrer, J.; Crausaz, F.; Riezman, H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J. Cell Biol. 1993, 120, 55–65. [Google Scholar]
- Chiou, J.C.; Li, X.P.; Remacha, M.; Ballesta, J.P.; Tumer, N.E. The ribosomal stalk is required for ribosome binding, depurination of the rRNA and cytotoxicity of ricin A chain in Saccharomyces cerevisiae. Mol. Microbiol. 2008, 70, 1441–1452. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Becker, B.; Schmitt, M.J. Adapting Yeast as Model to Study Ricin Toxin A Uptake and Trafficking. Toxins 2011, 3, 834-847. https://doi.org/10.3390/toxins3070834
Becker B, Schmitt MJ. Adapting Yeast as Model to Study Ricin Toxin A Uptake and Trafficking. Toxins. 2011; 3(7):834-847. https://doi.org/10.3390/toxins3070834
Chicago/Turabian StyleBecker, Björn, and Manfred J. Schmitt. 2011. "Adapting Yeast as Model to Study Ricin Toxin A Uptake and Trafficking" Toxins 3, no. 7: 834-847. https://doi.org/10.3390/toxins3070834
APA StyleBecker, B., & Schmitt, M. J. (2011). Adapting Yeast as Model to Study Ricin Toxin A Uptake and Trafficking. Toxins, 3(7), 834-847. https://doi.org/10.3390/toxins3070834




