A Conformational Shift in the Dissociated Cholera Toxin A1 Subunit Prevents Reassembly of the Cholera Holotoxin
Abstract
:1. Introduction

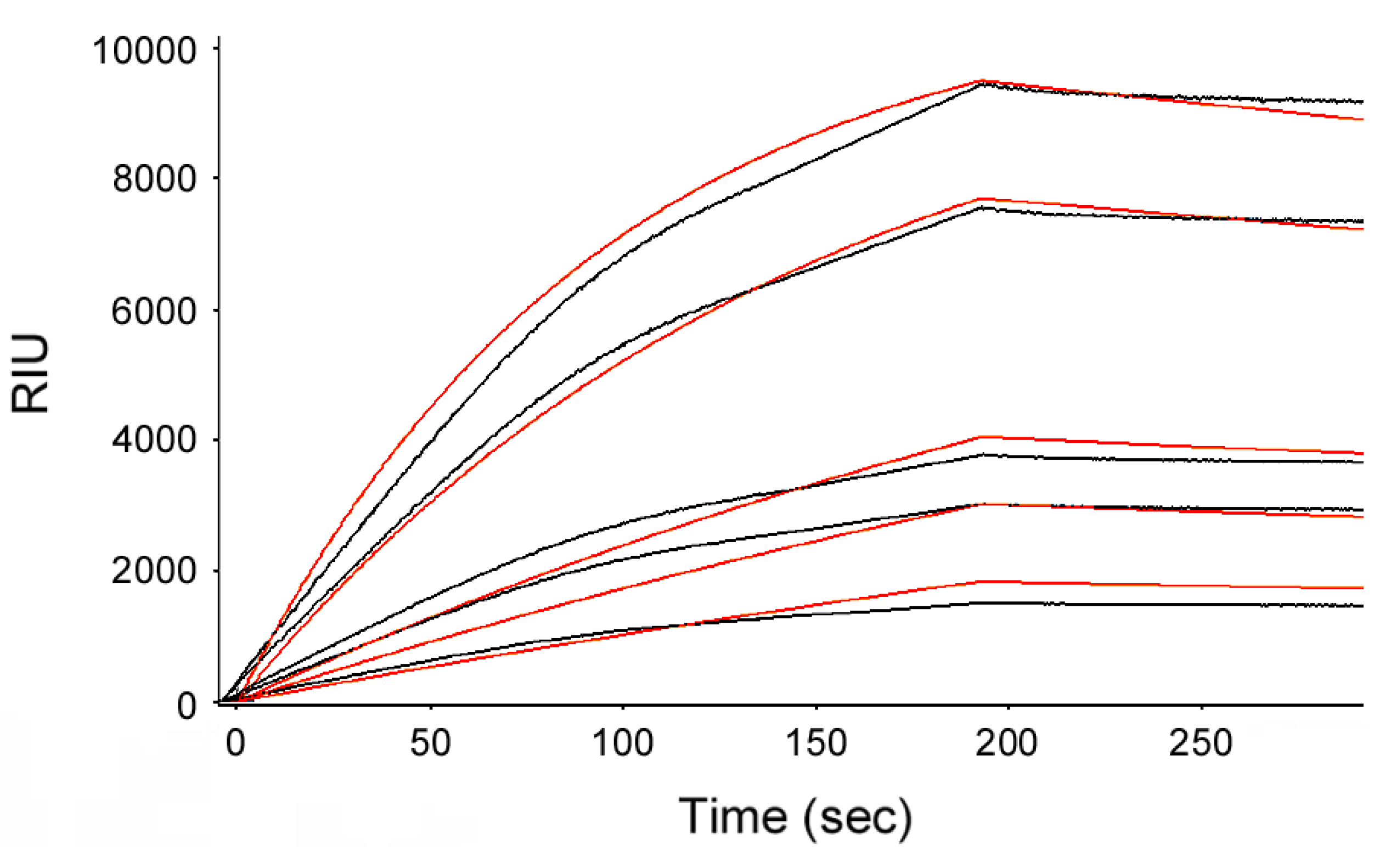
2. Results and Discussion



3. Experimental Section
3.1. Isolation of CTA1 from CTA
3.2. Holotoxin Assembly Detected by SPR
3.3. Intoxication Assay with Re-Assembled Holotoxin
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- De Haan, L.; Hirst, T.R. Cholera toxin: A paradigm for multi-functional engagement of cellular mechanisms (Review). Mol. Membr. Biol. 2004, 21, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, J.; Holmgren, J. Cholera toxin—A foe & a friend. Indian J. Med. Res. 2011, 133, 153–163. [Google Scholar] [PubMed]
- Hirst, T.R.; Holmgren, J. Conformation of protein secreted across bacterial outer membranes: A study of enterotoxin translocation from Vibrio cholerae. Proc. Natl. Acad. Sci. USA 1987, 84, 7418–7422. [Google Scholar] [CrossRef] [PubMed]
- Booth, B.A.; Boesman-Finkelstein, M.; Finkelstein, R.A. Vibrio cholerae hemagglutinin/protease nicks cholera enterotoxin. Infect. Immun. 1984, 45, 558–560. [Google Scholar] [PubMed]
- Mekalanos, J.J.; Collier, R.J.; Romig, W.R. Enzymic activity of cholera toxin. II. Relationships to proteolytic processing, disulfide bond reduction, and subunit composition. J. Biol. Chem. 1979, 254, 5855–5861. [Google Scholar] [PubMed]
- Lencer, W.I.; Constable, C.; Moe, S.; Jobling, M.G.; Webb, H.M.; Ruston, S.; Madara, J.L.; Hirst, T.R.; Holmes, R.K. Targeting of cholera toxin and Escherichia coli heat labile toxin in polarized epithelia: Role of COOH-terminal KDEL. J. Cell Biol. 1995, 131, 951–962. [Google Scholar] [CrossRef] [PubMed]
- O’Neal, C.J.; Amaya, E.I.; Jobling, M.G.; Holmes, R.K.; Hol, W.G. Crystal structures of an intrinsically active cholera toxin mutant yield insight into the toxin activation mechanism. Biochemistry 2004, 43, 3772–3782. [Google Scholar] [CrossRef] [PubMed]
- Wernick, N.L.B.; Chinnapen, D.J.-F.; Cho, J.A.; Lencer, W.I. Cholera toxin: An intracellular journey into the cytosol by way of the endoplasmic reticulum. Toxins 2010, 2, 310–325. [Google Scholar] [CrossRef] [PubMed]
- Pande, A.H.; Scaglione, P.; Taylor, M.; Nemec, K.N.; Tuthill, S.; Moe, D.; Holmes, R.K.; Tatulian, S.A.; Teter, K. Conformational instability of the cholera toxin A1 polypeptide. J. Mol. Biol. 2007, 374, 1114–1128. [Google Scholar] [CrossRef] [PubMed]
- Goins, B.; Freire, E. Thermal stability and intersubunit interactions of cholera toxin in solution and in association with its cell-surface receptor ganglioside GM1. Biochemistry 1988, 27, 2046–2052. [Google Scholar] [CrossRef] [PubMed]
- Surewicz, W.K.; Leddy, J.J.; Mantsch, H.H. Structure, stability, and receptor interaction of cholera toxin as studied by Fourier-transform infrared spectroscopy. Biochemistry 1990, 29, 8106–8111. [Google Scholar] [CrossRef] [PubMed]
- Majoul, I.; Ferrari, D.; Soling, H.D. Reduction of protein disulfide bonds in an oxidizing environment. The disulfide bridge of cholera toxin A-subunit is reduced in the endoplasmic reticulum. FEBS Lett. 1997, 401, 104–108. [Google Scholar] [CrossRef]
- Tomasi, M.; Battistini, A.; Araco, A.; Roda, L.G.; D’Agnolo, G. The role of the reactive disulfide bond in the interaction of cholera-toxin functional regions. Eur. J. Biochem. 1979, 93, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.; Burress, H.; Banerjee, T.; Ray, S.; Curtis, D.; Tatulian, S.A.; Teter, K. Substrate-induced unfolding of protein disulfide isomerase displaces the cholera toxin A1 subunit from its holotoxin. PLoS Pathog. 2014, 10, e1003925. [Google Scholar] [CrossRef] [PubMed]
- Tsai, B.; Rodighiero, C.; Lencer, W.I.; Rapoport, T.A. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 2001, 104, 937–948. [Google Scholar] [CrossRef]
- Taylor, M.; Banerjee, T.; Ray, S.; Tatulian, S.A.; Teter, K. Protein disulfide isomerase displaces the cholera toxin A1 subunit from the holotoxin without unfolding the A1 subunit. J. Biol. Chem. 2011, 286, 22090–22100. [Google Scholar] [CrossRef] [PubMed]
- Massey, S.; Banerjee, T.; Pande, A.H.; Taylor, M.; Tatulian, S.A.; Teter, K. Stabilization of the tertiary structure of the cholera toxin A1 subunit inhibits toxin dislocation and cellular intoxication. J. Mol. Biol. 2009, 393, 1083–1096. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.; Banerjee, T.; Navarro-Garcia, F.; Huerta, J.; Massey, S.; Burlingame, M.; Pande, A.H.; Tatulian, S.A.; Teter, K. A therapeutic chemical chaperone inhibits cholera intoxication and unfolding/translocation of the cholera toxin A1 subunit. PLoS ONE 2011, 6, e18825. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, T.; Pande, A.; Jobling, M.G.; Taylor, M.; Massey, S.; Holmes, R.K.; Tatulian, S.A.; Teter, K. Contribution of subdomain structure to the thermal stability of the cholera toxin A1 subunit. Biochemistry 2010, 49, 8839–8846. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, K.M.; Forster, M.L.; Lencer, W.I.; Tsai, B. Derlin-1 facilitates the retro-translocation of cholera toxin. Mol. Biol. Cell. 2008, 19, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, K.M.; Williams, J.M.; Kikkert, M.; van Voorden, S.; Wiertz, E.J.; Ye, Y.; Tsai, B. The E3 ubiquitin ligases Hrd1 and gp78 bind to and promote cholera toxin retro-translocation. Mol. Biol. Cell. 2010, 21, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Dixit, G.; Mikoryak, C.; Hayslett, T.; Bhat, A.; Draper, R.K. Cholera toxin up-regulates endoplasmic reticulum proteins that correlate with sensitivity to the toxin. Exp. Biol. Med. 2008, 233, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Saslowsky, D.E.; Cho, J.A.; Chinnapen, H.; Massol, R.H.; Chinnapen, D.J.; Wagner, J.S.; de Luca, H.E.; Kam, W.; Paw, B.H.; Lencer, W.I. Intoxication of zebrafish and mammalian cells by cholera toxin depends on the flotillin/reggie proteins but not Derlin-1 or -2. J. Clin. Investig. 2010, 120, 4399–4409. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, A.; Herrgen, H.; Winkeler, A.; Herzog, V. Cholera toxin is exported from microsomes by the Sec61p complex. J. Cell Biol. 2000, 148, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Teter, K.; Holmes, R.K. Inhibition of endoplasmic reticulum-associated degradation in CHO cells resistant to cholera toxin, Pseudomonas aeruginosa exotoxin A, and ricin. Infect. Immun. 2002, 70, 6172–6179. [Google Scholar] [CrossRef] [PubMed]
- Teter, K. Toxin instability and its role in toxin translocation from the endoplasmic reticulum to the cytosol. Biomolecules 2013, 3, 997–1029. [Google Scholar] [CrossRef] [PubMed]
- Hazes, B.; Read, R.J. Accumulating evidence suggests that several AB-toxins subvert the endoplasmic reticulum-associated protein degradation pathway to enter target cells. Biochemistry 1997, 36, 11051–11054. [Google Scholar] [CrossRef] [PubMed]
- Rodighiero, C.; Tsai, B.; Rapoport, T.A.; Lencer, W.I. Role of ubiquitination in retro-translocation of cholera toxin and escape of cytosolic degradation. EMBO Rep. 2002, 3, 1222–1227. [Google Scholar] [CrossRef] [PubMed]
- Murayama, T.; Tsai, S.C.; Adamik, R.; Moss, J.; Vaughan, M. Effects of temperature on ADP-ribosylation factor stimulation of cholera toxin activity. Biochemistry 1993, 32, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Ampapathi, R.S.; Creath, A.L.; Lou, D.I.; Craft, J.W., Jr.; Blanke, S.R.; Legge, G.B. Order-disorder-order transitions mediate the activation of cholera toxin. J. Mol. Biol. 2008, 377, 748–760. [Google Scholar] [CrossRef] [PubMed]
- Welsh, C.F.; Moss, J.; Vaughan, M. ADP-ribosylation factors: A family of approximately 20-kDa guanine nucleotide-binding proteins that activate cholera toxin. Mol. Cell. Biochem. 1994, 138, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Taylor, M.; Banerjee, T.; Tatulian, S.A.; Teter, K. Lipid rafts alter the stability and activity of the cholera toxin A1 subunit. J. Biol. Chem. 2012, 287, 30395–30405. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, T.; Taylor, M.; Jobling, M.G.; Burress, H.; Yang, Z.; Serrano, A.; Holmes, R.K.; Tatulian, S.A.; Teter, K. ADP-ribosylation factor 6 acts as an allosteric activator for the folded but not disordered cholera toxin A1 polypeptide. Mol. Microbiol. 2014, 94, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Burress, H.; Taylor, M.; Banerjee, T.; Tatulian, S.A.; Teter, K. Co- and post-translocation roles for Hsp90 in cholera intoxication. J. Biol. Chem. 2014, 289, 33644–33654. [Google Scholar] [CrossRef] [PubMed]
- Spooner, R.A.; Watson, P.D.; Marsden, C.J.; Smith, D.C.; Moore, K.A.; Cook, J.P.; Lord, J.M.; Roberts, L.M. Protein disulphide-isomerase reduces ricin to its A and B chains in the endoplasmic reticulum. Biochem. J. 2004, 383, 285–293. [Google Scholar] [PubMed]
- Simpson, J.C.; Roberts, L.M.; Lord, J.M. Catalytic and cytotoxic activities of recombinant ricin A chain mutants with charged residues added at the carboxyl terminus. Protein Expr. Purif. 1995, 6, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Homola, J. Present and future of surface plasmon resonance biosensors. Anal. Bioanal. Chem. 2003, 377, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Medaglia, M.V.; Fisher, R.J. Analysis of Interacting Proteins with Surface Plasmon Resonance Using Biacore. In Protein-Protein Interactions; Golemis, E., Ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2002; pp. 255–272. [Google Scholar]
- Willander, M.; Al-Hilli, S. Analysis of biomolecules using surface plasmons. Methods Mol. Biol. 2009, 544, 201–229. [Google Scholar] [PubMed]
- Mekalanos, J.J.; Collier, R.J.; Romig, W.R. Purification of cholera toxin and its subunits: New methods of preparation and the use of hypertoxinogenic mutants. Infect. Immun. 1978, 20, 552–558. [Google Scholar] [PubMed]
- Holmes, R.K.; Twiddy, E.M. Characterization of monoclonal antibodies that react with unique and cross-reacting determinants of cholera enterotoxin and its subunits. Infect. Immun. 1983, 42, 914–923. [Google Scholar] [PubMed]
- Norton, E.B.; Lawson, L.B.; Mahdi, Z.; Freytag, L.C.; Clements, J.D. The A Subunit of Escherichia coli Heat-Labile Enterotoxin Functions as a Mucosal Adjuvant and Promotes IgG2a, IgA, and Th17 Responses to Vaccine Antigens. Infect. Immun. 2012, 80, 2426–2435. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Ryu, S.H.; Lee, S.H.; Lee, Y.H.; Lee, S.R.; Huh, J.W.; Kim, S.U.; Kim, E.; Kim, S.; Jon, S.; et al. Instability of toxin A subunit of AB5 toxins in the bacterial periplasm caused by deficiency of their cognate B subunits. Biochim. Biophys. Acta 2011, 1808, 2359–2365. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taylor, M.; Curtis, D.; Teter, K. A Conformational Shift in the Dissociated Cholera Toxin A1 Subunit Prevents Reassembly of the Cholera Holotoxin. Toxins 2015, 7, 2674-2684. https://doi.org/10.3390/toxins7072674
Taylor M, Curtis D, Teter K. A Conformational Shift in the Dissociated Cholera Toxin A1 Subunit Prevents Reassembly of the Cholera Holotoxin. Toxins. 2015; 7(7):2674-2684. https://doi.org/10.3390/toxins7072674
Chicago/Turabian StyleTaylor, Michael, David Curtis, and Ken Teter. 2015. "A Conformational Shift in the Dissociated Cholera Toxin A1 Subunit Prevents Reassembly of the Cholera Holotoxin" Toxins 7, no. 7: 2674-2684. https://doi.org/10.3390/toxins7072674




