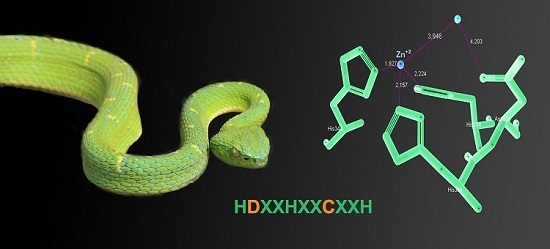
Novel Catalytically-Inactive PII Metalloproteinases from a Viperid Snake Venom with Substitutions in the Canonical Zinc-Binding Motif
Abstract
:1. Introduction
2. Results
2.1. Purification of SVMP BlatPII
2.2. Determination of Internal Peptide Sequences
2.3. Cloning of a P-II SVMP
2.4. Proteolytic Activity on Azocasein and Gelatin
2.5. Hemorrhagic, Edema-Forming, Coagulant and Platelet Aggregation Inhibitory Activities
2.6. Three-Dimensional Model of the Active Site of BlatPII-c
3. Discussion
4. Materials and Methods
4.1. Venom and Toxins
4.2. Purification of SVMP
4.3. HPLC Separation of Isoforms
4.4. Mass Spectrometry Analyses
4.4.1. MALDI-TOF/TOF
4.4.2. nLC-MS/MS
4.5. Determination of Internal Peptide Sequences by Edman Degradation
4.6. Detection of Carbohydrates
4.7. PCR Amplification of SVMPs cDNA
4.8. Proteolytic Activity on Azocasein
4.9. Proteolytic Activity on Gelatin
4.10. Local and Systemic Hemorrhagic Activities
4.11. Edema-Forming Activity
4.12. Coagulant Activity
4.13. Platelet Aggregation Inhibitory Activity
4.14. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fox, J.W.; Serrano, S.M.T. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon 2005, 45, 969–985. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Rucavado, A.; Escalante, T. Snake Venom Metalloproteinases. Biological Roles and Participation in the Pathophysiology of Envenomation. In Handbook of Venoms and Toxins of Reptiles; Mackessy, S.P., Ed.; CRC Press: Boca Ratón, FL, USA, 2010; pp. 115–138. [Google Scholar]
- Calvete, J.J. Proteomic tools against the neglected pathology of snake bite envenoming. Expert Rev. Proteom. 2011, 8, 739–758. [Google Scholar] [CrossRef] [PubMed]
- Bode, W.; Grams, F.; Reinemer, P.; Gomis-Rüth, F.-X.; Baumann, U.; McKay, D.; Stöcker, W. The Metzincin-superfamily of zinc-peptidases. Intracell. Protein Catabolism 1996, 389, 1–11. [Google Scholar]
- Fox, J.W.; Serrano, S.M. Snake venom metalloproteinases. In Handbook of Venoms and Toxins of Reptiles; Mackessy, S.P., Ed.; CRP Press: Boca Ratón, FL, USA, 2009; pp. 95–113. [Google Scholar]
- Casewell, N.R.; Sunagar, K.; Takacs, Z.; Calvete, J.J.; Jacson, T.N.W.; Fry, B.G. Snake Venom Metalloprotease Enzymes. In Venomous, Reptiles and Their Toxins. Evolution, Pathophysiology and Biodiscovery; Fry, B.G., Ed.; Oxford University Press: Oxford, UK, 2015; pp. 347–363. [Google Scholar]
- Tanjoni, I.; Evangelista, K.; Della-Casa, M.S.; Butera, D.; Magalhães, G.S.; Baldo, C.; Clissa, P.B.; Fernandes, I.; Eble, J.; Moura-da-Silva, A.M. Different regions of the class P-III snake venom metalloproteinase jararhagin are involved in binding to alpha2beta1 integrin and collagen. Toxicon 2010, 55, 1093–1099. [Google Scholar] [CrossRef] [PubMed]
- Moura-da-Silva, A.M.; Serrano, S.M.; Fox, J.W.; Gutierrez, J.M. Snake Venom Metalloproteinases. Structure, function and effects on snake bite pathology. In Animal Toxins: State of the Art. Perspectives in Health and Biotechonology; de Lima, M.E., Monteiro de Castro, A., Martin-Euclaire, M.F., Benedeta, R., Eds.; UFMG: Belo Horizonte, Brasil, 2009; pp. 525–546. [Google Scholar]
- Casewell, N.R.; Wagstaff, S.C.; Harrison, R.A.; Renjifo, C.; Wüster, W. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol. Biol. Evol. 2011, 28, 2637–2649. [Google Scholar] [CrossRef] [PubMed]
- Moura-da-Silva, A.M.; Theakston, R.D.G.; Crampton, J.M. Evolution of disintegrin cysteine-rich and mammalian matrix-degrading metalloproteinases: Gene duplication and divergence of a common ancestor rather than convergent evolution. J. Mol. Evol. 1996, 43, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Fry, B. From genome to “venome”: Molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005, 15, 403–420. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R. On the ancestral recruitment of metalloproteinases into the venom of snakes. Toxicon 2012, 60, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Juarez, P.; Comas, I.; Gonzalez-Candelas, F.; Calvete, J.J. Evolution of snake venom disintegrins by positive Darwinian selection. Mol. Biol. Evol. 2008, 25, 2391–2407. [Google Scholar] [CrossRef] [PubMed]
- Solórzano, A. Snakes of Costa Rica Distribution, Taxonomy, and Natural History; INBio: Heredia, Costa Rica, 2004. [Google Scholar]
- Carbajo, R.J.; Sanz, L.; Perez, A.; Calvete, J.J. NMR structure of bitistatin—A missing piece in the evolutionary pathway of snake venom disintegrins. FEBS J. 2015, 282, 341–360. [Google Scholar] [CrossRef] [PubMed]
- Serrano, S.M.T.; Jia, L.-G.; Wang, D.; Shannon, J.D.; Fox, J.W. Function of the cysteine-rich domain of the haemorrhagic metalloproteinase atrolysin A: Targeting adhesion proteins collagen I and von Willebrand factor. Biochem. J. 2005, 391, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative protein structure modeling using MODELLER. In Current Protocols in Bioinformatics; John Wiley & Sons, Inc: Hoboken, NJ, USA, 2002; Volume 47, pp. 5.6.1–5.6.32. [Google Scholar]
- Lovejoy, B.; Hassell, A.M.; Luther, M.A.; Weigl, D.; Jordan, S.R. Crystal Structures of Recombinant 19-kDa Human Fibroblast Collagenase Complexed to Itself. Biochemistry 1994, 33, 8207–8217. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, M.; Floyd, C.D.; Brown, P.; Gearing, A.J.H. Design and Therapeutic Application of Matrix Metalloproteinase Inhibitors. Chem. Rev. 1999, 99, 2735–2776. [Google Scholar] [CrossRef] [PubMed]
- Bates, E.E.M.; Fridman, W.H.; Mueller, C.G.F. The ADAMDEC1 (decysin) gene structure: Evolution by duplication in a metalloprotease gene cluster on chromosome 8p12. Immunogenetics 2002, 54, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Edwards, D.R.; Handsley, M.M.; Pennington, C.J. The ADAM metalloproteinases. Mol. Aspects Med. 2008, 29, 258–89. [Google Scholar] [CrossRef] [PubMed]
- Frayne, J.A.N.; Jury, J.A.; Barker, H.L.; Hall, L.E.N. Rat MDC Family of Proteins: Sequence Analysis, Tissue Distribution, and Expression in Prepubertal. Mol. Reprod. Dev. 1997, 167, 159–167. [Google Scholar] [CrossRef]
- Liu, H.; Shim, A.H.R.; He, X. Structural characterization of the ectodomain of a disintegrin and metalloproteinase-22 (ADAM22), a neural adhesion receptor instead of metalloproteinase: Insights on ADAM function. J. Biol. Chem. 2009, 284, 29077–29086. [Google Scholar] [CrossRef] [PubMed]
- Waters, S.I.; White, J.M. Biochemical and Molecular Characterization of Bovine Fertilin a and (ADAM 1 and ADAM 2): A Candidate Sperm-Egg Binding/Fusion Complex. Biol. Reprod. 1997, 56, 1245–1254. [Google Scholar] [CrossRef] [PubMed]
- Cornwall, G.A.; Hsia, N. ADAM7, A Member of the ADAM (A Disintegrin And Metalloprotease) Gene Family is Specifically Expressed in the Mouse Anterior Pituitary and Epididymis. Endocrinology 1997, 138, 4262–4272. [Google Scholar] [CrossRef] [PubMed]
- Petretski, J.H.; Kanashiro, M.M.; Rodrigues, F.R.; Alves, E.W.; Machado, O.L.; Kipnis, T.L. Edema induction by the disintegrin-like/cysteine-rich domains from a Bothrops atrox hemorrhagin. Biochem. Biophys. Res. Commun. 2000, 276, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Kamiguti, A.S.; Zuzel, M.; Reid, A. Snake venom metalloproteinases and disintegrins: Interactions with cells. Braz. J. Med. Biol. Res. 1998, 31, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Marcinkiewicz, C.; Monleón, D.; Esteve, V.; Celda, B.; Juárez, P.; Sanz, L. Snake venom disintegrins: Evolution of structure and function. Toxicon 2005, 45, 1063–1074. [Google Scholar] [CrossRef] [PubMed]
- Wijeyewickrema, L.C.; Berndt, M.C.; Andrews, R.K. Snake venom probes of platelet adhesion receptors and their ligands. Toxicon 2005, 45, 1051–1061. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Rangel, J. Snake venom Lys49 myotoxins: From phospholipases A2 to non-enzymatic membrane disruptors. Toxicon 2012, 60, 520–530. [Google Scholar] [CrossRef] [PubMed]
- Kurtović, T.; Brgles, M.; Leonardi, A.; Lang Balija, M.; Sajevic, T.; Križaj, I.; Allmaier, G.; Marchetti-Deschmann, M.; Halassy, B. VaSP1, catalytically active serine proteinase from Vipera ammodytes ammodytes venom with unconventional active site triad. Toxicon 2014, 77, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Jin, Y.; Zhong, S.; Chen, R.; Zhu, S.; Wang, W.; Lu, Q.; Xiong, Y. A unique group of inactive serine protease homologues from snake venom. Toxicon 2008, 52, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Sanz, L.; Angulo, Y.; Calvete, J.J.; Escolano, J.; Fernández, J.; Sanz, L.; Angulo, Y.; Gutiérrez, J.M.; Calvete, J.J. Snake venomics and antivenomics of the arboreal neotropical pitvipers Bothriechis lateralis and Bothriechis schlegelii. J. Proteome Res. 2008, 7, 2445–2457. [Google Scholar] [CrossRef] [PubMed]
- Camacho, E.; Villalobos, E.; Sanz, L.; Pérez, A.; Escalante, T.; Lomonte, B.; Calvete, J.J.; Gutiérrez, J.M.; Rucavado, A. Understanding structural and functional aspects of PII snake venom metalloproteinases: Characterization of BlatH1, a hemorrhagic dimeric enzyme from the venom of Bothriechis lateralis. Biochimie 2014, 101, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Romero, M.; Diaz, C.; Borkow, G.A.D.; Ovadia, M. Isolation and characterization of a metalloproteinase with weak activity from the venom of the snake Bothrops asper (Terciopelo). Toxicon 1995, 33, 19–29. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Leprevost, F.V.; Valente, R.H.; Lima, D.B.; Perales, J.; Melani, R.; Iii, J.R.Y.; Barbosa, V.C.; Junqueira, M.; Carvalho, P.C. PepExplorer: A Similarity-driven Tool for Analyzing de Novo Sequencing Results. Mol. Cell. Proteom. 2014, 13, 2480–2489. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Shih, C.; Huang, T. A novel P-I class metalloproteinase with broad substrate-cleaving activity, agkislysin, from Agkistrodon acutus venom. Biochem. Biophys. Res. Commun. 2004, 324, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Herron, G.S.; Banda, M.J.; Clark, E.J.; Gavrilovic, J.; Werb, Z. Secretion of Metalloproteinases by Stimulated Capillary Endothelial Cells. J. Biol. Chem. 1986, 261, 2814–2818. [Google Scholar] [PubMed]
- Rucavado, A.; Núñez, J.; Gutiérrez, J.M. Blister formation and skin damage induced by BaP1, a haemorrhagic metalloproteinase from the venom of the snake Bothrops asper. J. Exp. Pathol. Int. 1998, 79, 245–254. [Google Scholar]
- Gutiérrez, J.; Gené, J.; Rojas, G.; Cerdas, L. Neutralization of proteolytic and hemorrhagic activities of Costa Rican snake venoms by a polyvalent antivenom. Toxicon 1985, 23, 887–893. [Google Scholar] [CrossRef]
- Escalante, T.; Núñez, J.; Moura da Silva, A.M.; Rucavado, A.; Theakston, R.D.G.; Gutiérrez, J.M. Pulmonary hemorrhage induced by jararhagin, a metalloproteinase from Bothrops jararaca snake venom. Toxicol. Appl. Pharmacol. 2003, 193, 17–28. [Google Scholar] [CrossRef]
- Lomonte, B.; Tarkowski, A.; Hanson, L.Å. Host response to Bothrops asper snake venom: Analysis of edema formation, inflammatory cells and cytokine release in a mouse model. Inflammation 1993, 17, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Gené, J.A.; Rojas, G.; Gutiérrez, J.M.; Cerdas, L. Comparative study on the coagulant, defibrinating, fibrinolytic and fibrinogenolytic activities of Costa Rican crotaline snake venoms and their neutralization by a polyvalent antivenom. Toxicon 1989, 27, 841–848. [Google Scholar] [CrossRef]
- Kamiguti, A.; Hay, C.; Zuzel, M. Inhibition of collagen-induced platelet aggregation as the result of cleavage of alpha 2 beta 1-integrin by the snake venom metalloproteinase jararhagin. Biochem. J. 1996, 320, 635–641. [Google Scholar] [CrossRef] [PubMed]

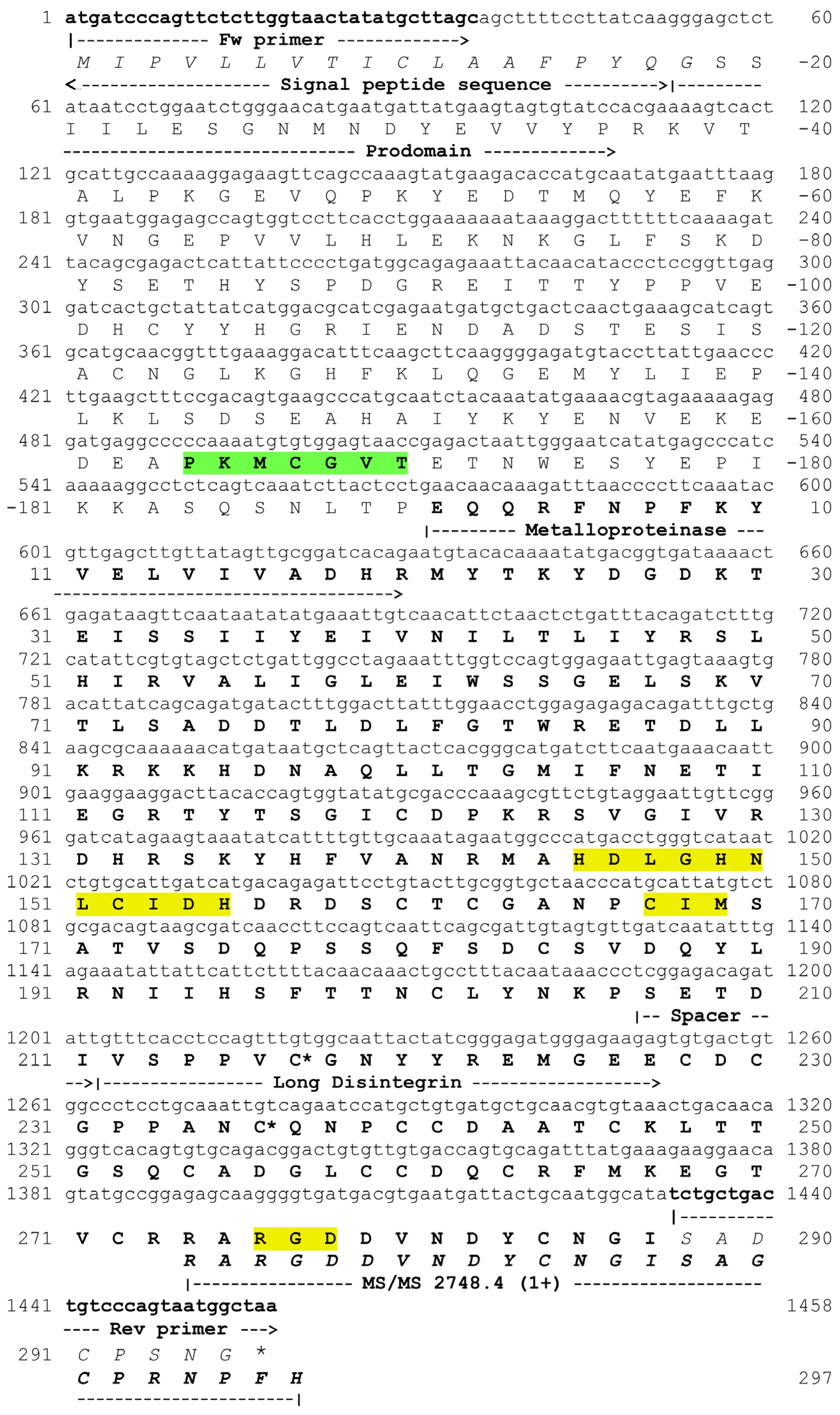




| Peptide Sequence | HPLC Peak | m/z | z | Confidence a | Error b (ppm) | Method c | Domain |
|---|---|---|---|---|---|---|---|
| YIELVIVADHR | 1 | 1327.8 | 1 | MV | 47.4 | MALDI/DE NOVO | metalloproteinase |
| NLTPEEQRY | 1 | 1149.6 | 1 | MV | 40.0 | MALDI/DE NOVO | metalloproteinase |
| SRYHFVANR | 1 | 1149.6 | 1 | MV | 7.8 | MALDI/DE NOVO | metalloproteinase |
| YHFVANR | 1 | - | MV | - | EDMAN | metalloproteinase | |
| MAHELGHNLGIHQDR | 1 | - | - | MV | - | EDMAN | metalloproteinase |
| MAHELGHNLGIHQDR | 1 | 1727.8 | 1 | MV | 22.5 | MALDI/DE NOVO | metalloproteinase |
| MAHELGHNLGLHQDR | 1 | 576.6180 | 3 | 99 | 0.2 | nESI/DE NOVO | metalloproteinase |
| DSCSCGSNSCIMSATVSNEPSSR | 1 | 2493.0 | 1 | MV | 10.0 | MALDI/DE NOVO | metalloproteinase |
| CIDNEPLR | 1 | 1016.5 | 1 | MV | 16.7 | MALDI/DE NOVO | metalloproteinase |
| NEPLRTDIVSPPFCGNYYPE * | 1 | 2368.3 | 1 | MV | 88.2 | MALDI/DE NOVO | metalloproteinase/disintegrin |
| LTTGSQCAEGLCCDQCR | 1 | 2015.9 | 1 | MV | 47.6 | MALDI/DE NOVO | disintegrin |
| FEGLCCDQCR | 1 | 1344.4 | 1 | MV | 84.0 | MALDI/DE NOVO | disintegrin |
| RTDIVSPPF ** | 1 | 1031.6 | 1 | MV | 46.5 | MALDI/DE NOVO | disintegrin |
| YIELVIVADHR | 2 | 1327.8 | 1 | MV | 47.4 | MALDI/DE NOVO | metalloproteinase |
| VTLSADDTLDLFGTWR | 2 | - | MV | - | EDMAN | metalloproteinase | |
| VALIGLEIWSSGELSK | 2 | 1702.0 | 1 | MV | 34.0 | MALDI/DE NOVO | metalloproteinase |
| YHFVANR | 2 | 906.5 | 1 | MV | 46.3 | MALDI/DE NOVO | metalloproteinase |
| MAHDLGHNLCIDHDR | 2 | 1803.9 | 1 | MV | 54.8 | MALDI/DE NOVO | metalloproteinase |
| MAHDLGHNLCLDHDR | 2 | 902.4039 | 2 | 99 | −0.4 | nESI/DE NOVO | metalloproteinase |
| MAHDLGHNLCLDHDDR | 2 | 959.9163 | 2 | 99 | −1.5 | nESI/DE NOVO | metalloproteinase |
| MAHELGHNLGLHQDDR | 2 | 614.9611 | 3 | 99 | 1.2 | nESI/DE NOVO | metalloproteinase |
| MAHELGHNLGLHQDR | 2 | 864.4214 | 2 | 99 | −2.1 | nESI/DE NOVO | metalloproteinase |
| NKPSETDIVSPPVCGNYY ** | 2 | 2040.1 | 1 | MV | 79.4 | MALDI/DE NOVO | metalloproteinase/disintegrin |
| CNGISAGCPRNPF ** | 2 | 1449.7 | 1 | MV | 44.1 | MALDI/DE NOVO | disintegrin |
| RARGDDVNDYCNGISAGCPRNPFH * | 2 | 2748.4 | 1 | MV | 68.7 | MALDI/DE NOVO | disintegrin |
| ARGDDVNDYCNGISAGCPR | 2 | 2097.1 | 1 | MV | 101.5 | MALDI/DE NOVO | disintegrin |
| ARGDDVDDYCDGLDAGCPR | 2 | 709.6204 | 3 | 91 | −1.7 | nESI/DE NOVO | disintegrin |
| AATCKLTTGSQCADGLCCDQCKFMRE * | 2 | 3068.5 | 1 | MV | 71.0 | MALDI/DE NOVO | disintegrin |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Camacho, E.; Sanz, L.; Escalante, T.; Pérez, A.; Villalta, F.; Lomonte, B.; Neves-Ferreira, A.G.C.; Feoli, A.; Calvete, J.J.; Gutiérrez, J.M.; et al. Novel Catalytically-Inactive PII Metalloproteinases from a Viperid Snake Venom with Substitutions in the Canonical Zinc-Binding Motif. Toxins 2016, 8, 292. https://doi.org/10.3390/toxins8100292
Camacho E, Sanz L, Escalante T, Pérez A, Villalta F, Lomonte B, Neves-Ferreira AGC, Feoli A, Calvete JJ, Gutiérrez JM, et al. Novel Catalytically-Inactive PII Metalloproteinases from a Viperid Snake Venom with Substitutions in the Canonical Zinc-Binding Motif. Toxins. 2016; 8(10):292. https://doi.org/10.3390/toxins8100292
Chicago/Turabian StyleCamacho, Erika, Libia Sanz, Teresa Escalante, Alicia Pérez, Fabián Villalta, Bruno Lomonte, Ana Gisele C. Neves-Ferreira, Andrés Feoli, Juan J. Calvete, José María Gutiérrez, and et al. 2016. "Novel Catalytically-Inactive PII Metalloproteinases from a Viperid Snake Venom with Substitutions in the Canonical Zinc-Binding Motif" Toxins 8, no. 10: 292. https://doi.org/10.3390/toxins8100292
APA StyleCamacho, E., Sanz, L., Escalante, T., Pérez, A., Villalta, F., Lomonte, B., Neves-Ferreira, A. G. C., Feoli, A., Calvete, J. J., Gutiérrez, J. M., & Rucavado, A. (2016). Novel Catalytically-Inactive PII Metalloproteinases from a Viperid Snake Venom with Substitutions in the Canonical Zinc-Binding Motif. Toxins, 8(10), 292. https://doi.org/10.3390/toxins8100292