Venom Gland Transcriptomic and Proteomic Analyses of the Enigmatic Scorpion Superstitionia donensis (Scorpiones: Superstitioniidae), with Insights on the Evolution of Its Venom Components
Abstract
:1. Introduction
Calcins, Scorpines, La1-Like and Potassium Channel κ Toxins in Scorpion Venoms
2. Results and Discussion
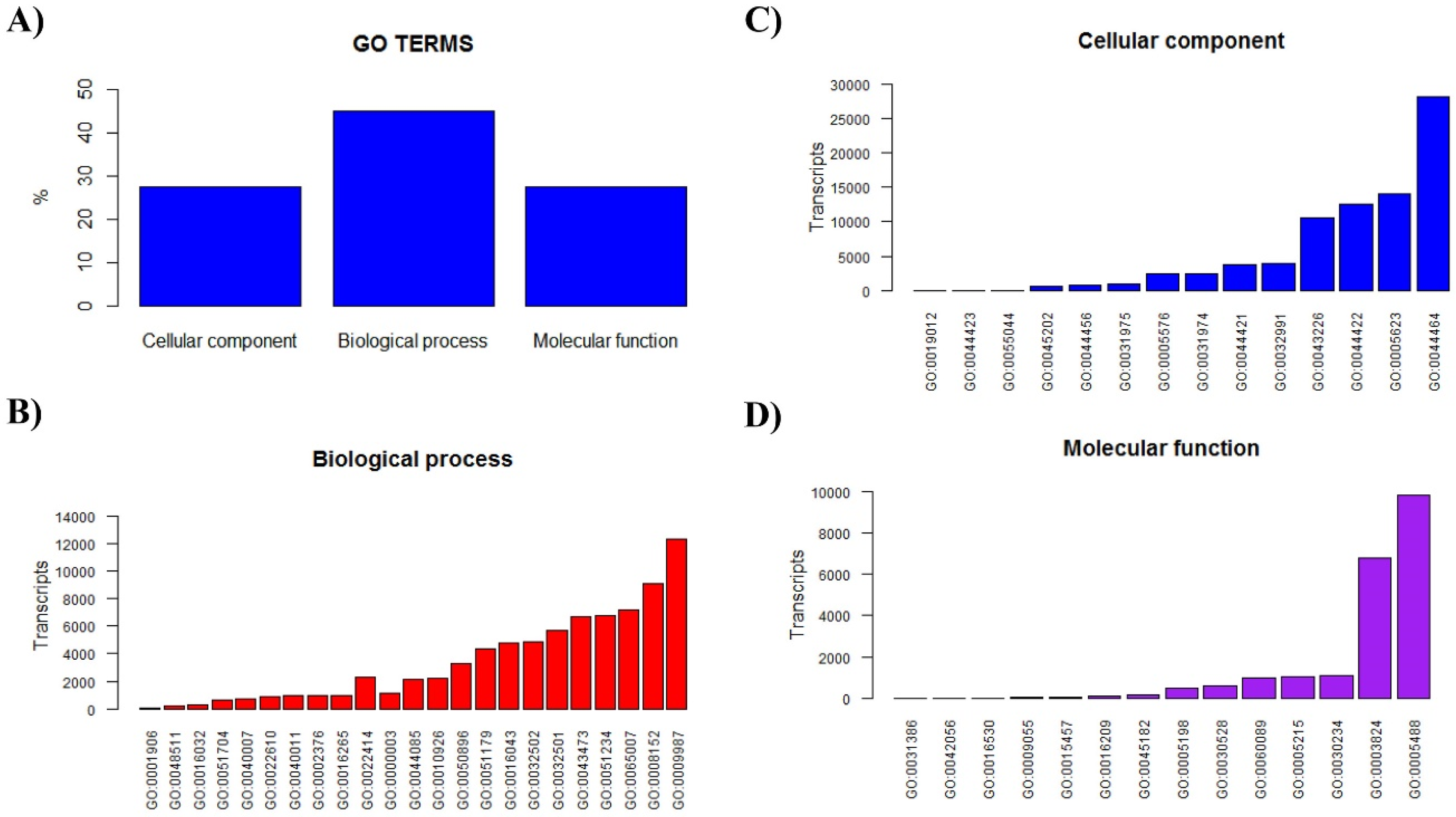
2.1. S. donensis Venom Gland Global Transcriptomic Analysis
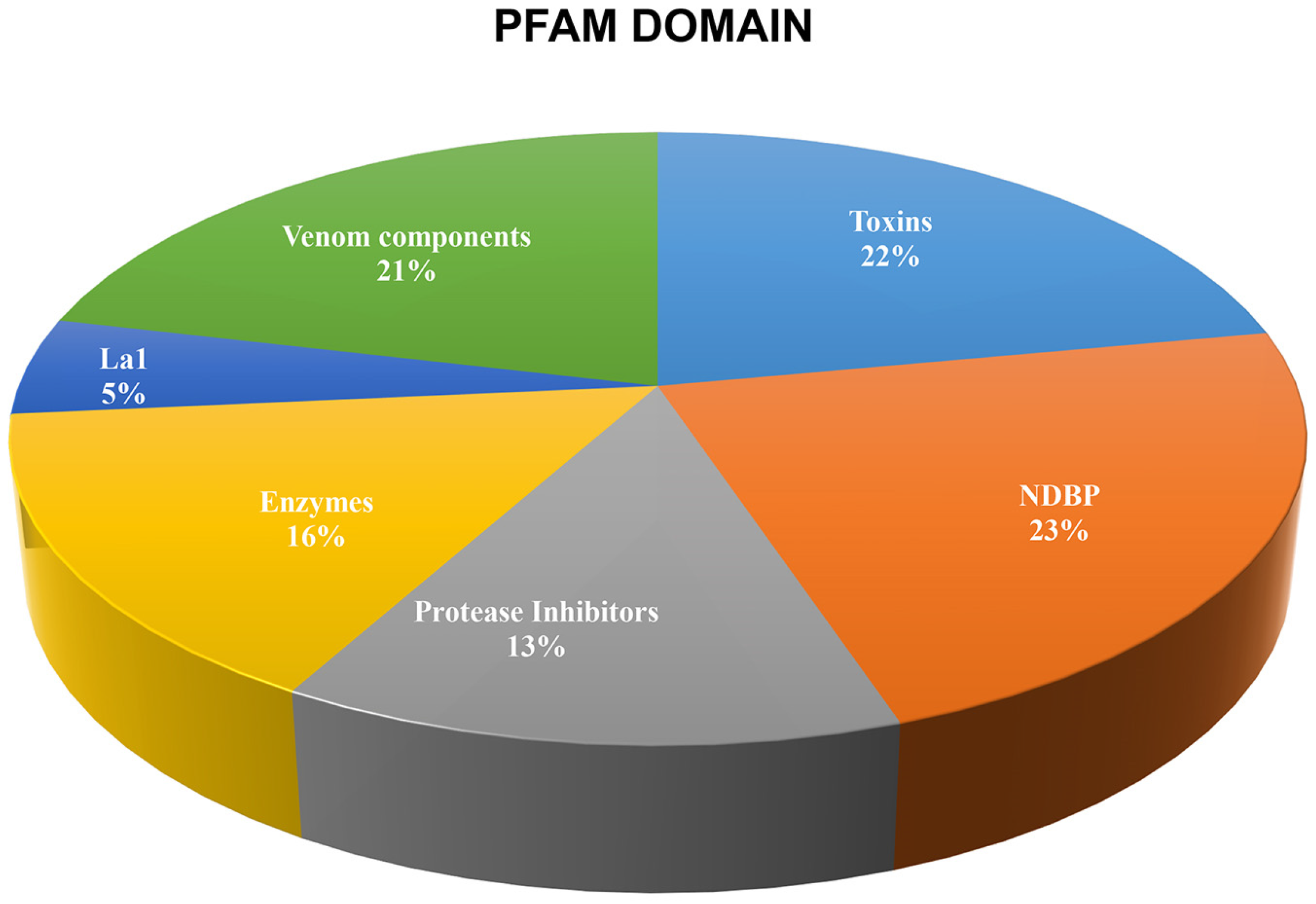
2.2. The Repertoire of Venom-Specific Transcripts in S. donensis
2.2.1. Toxins
Sodium Channel Toxins
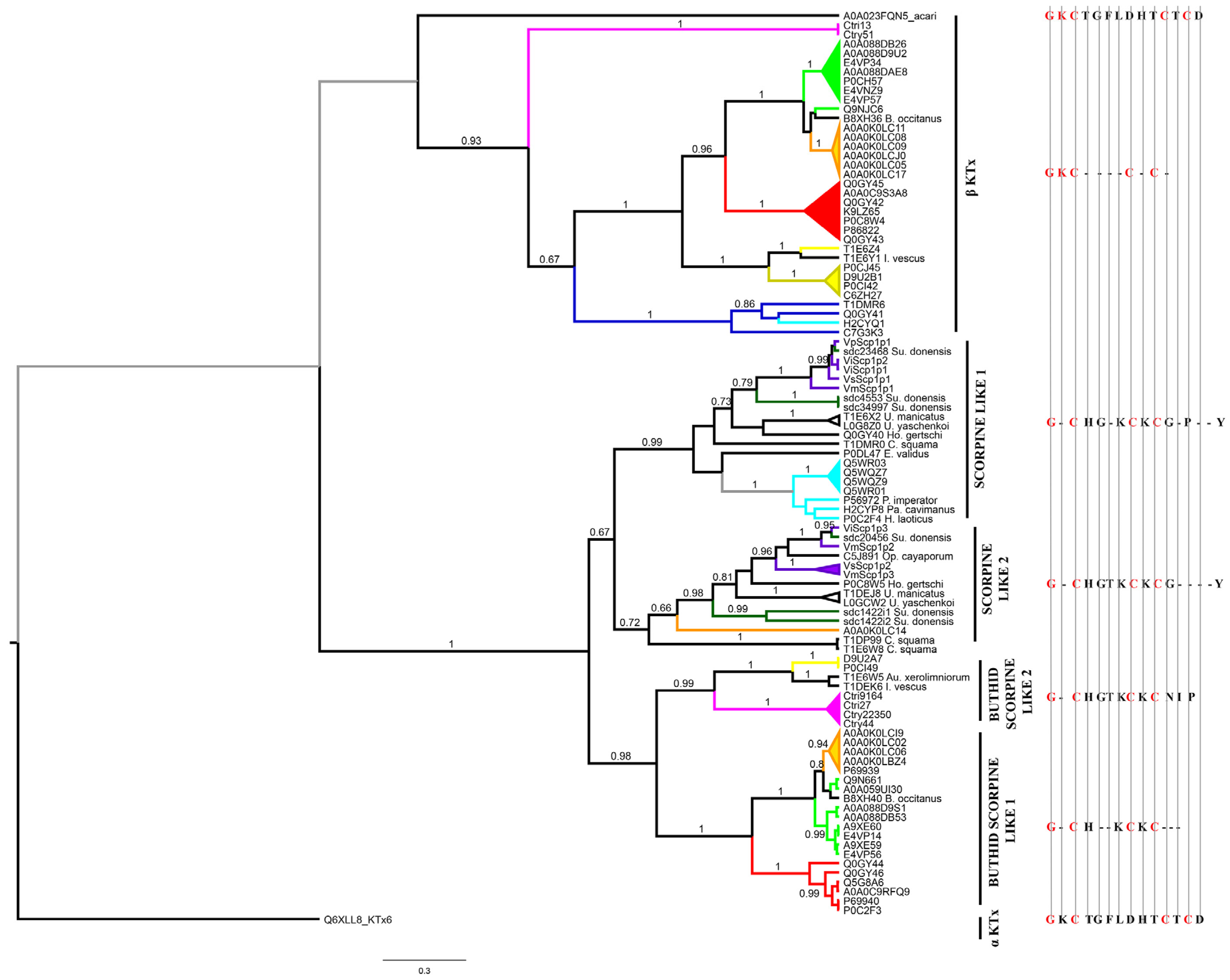
Potassium channel toxins
Scorpine-like peptides
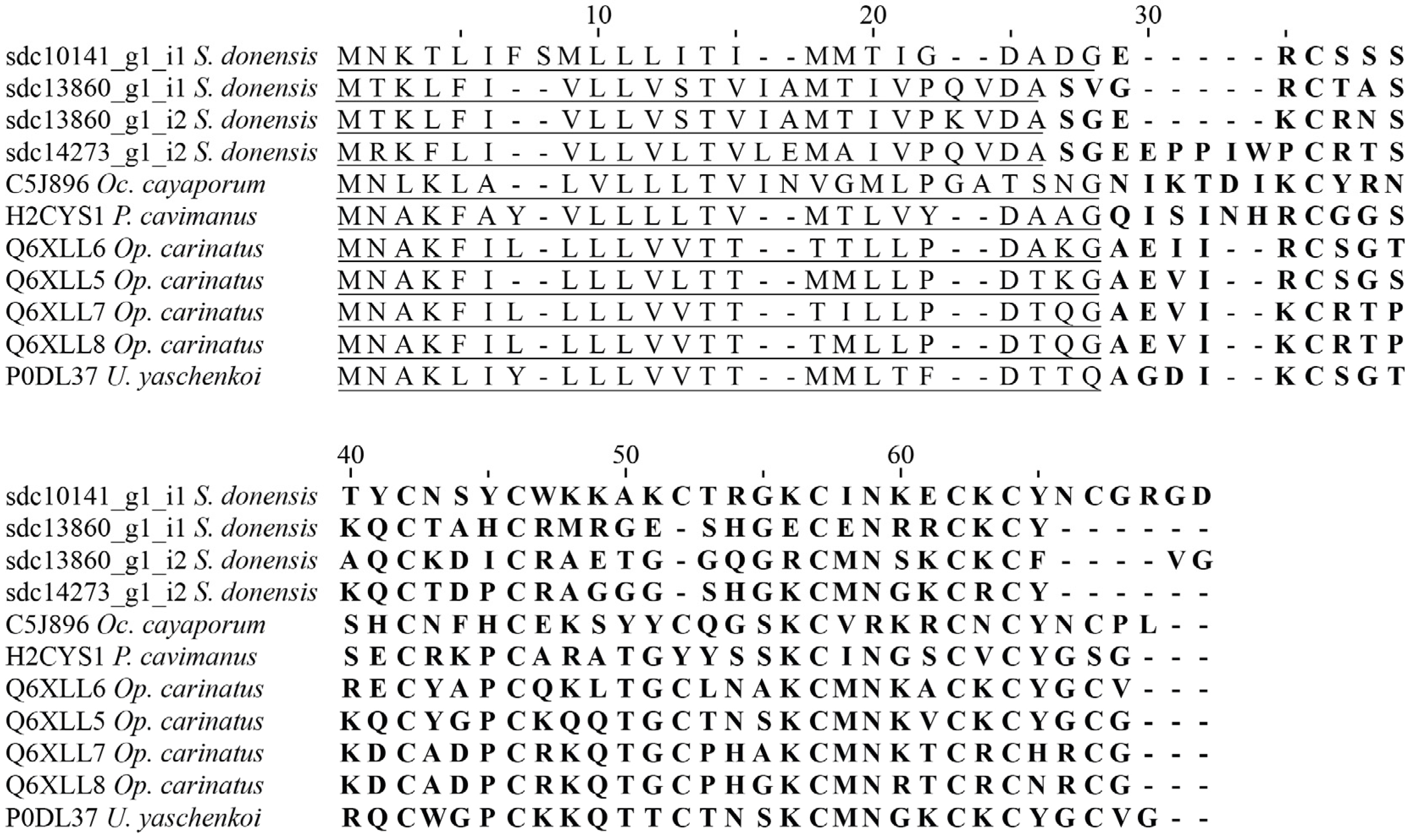
Calcins
2.2.2. Non-Disulfide-Bridged Peptides (NDBPs)
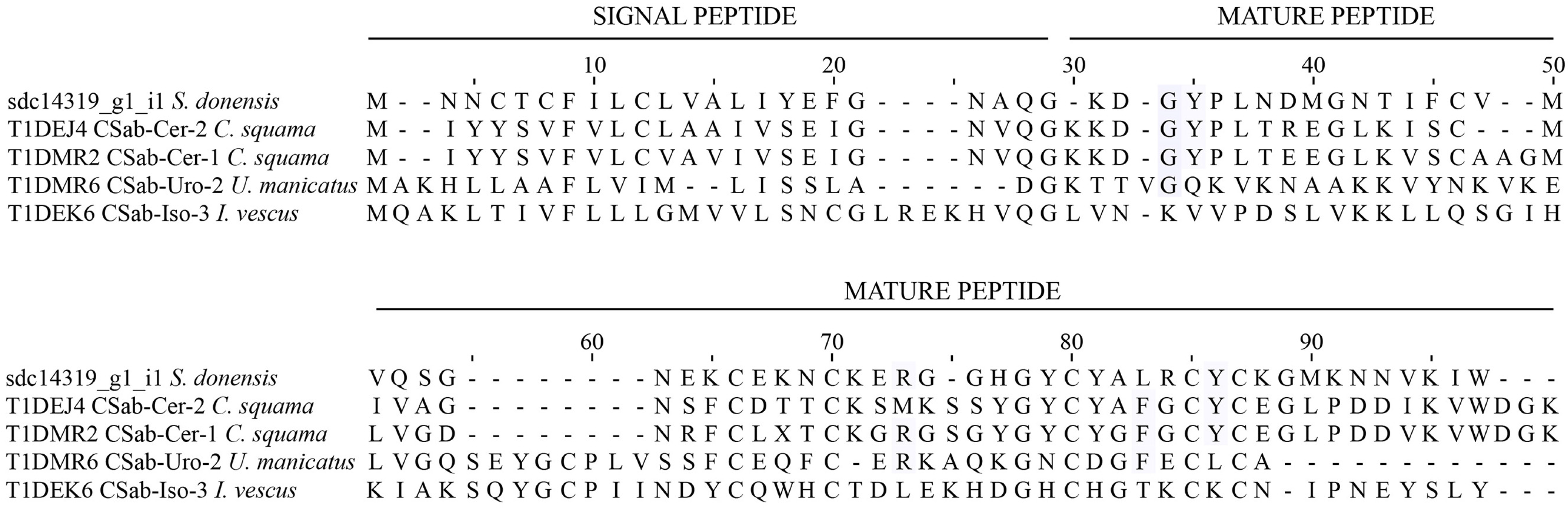
2.2.3. La1-Like Peptides
2.2.4. Enzymes
2.2.5. Protease Inhibitors
2.2.6. Other Venom Components
2.3. Amino Acid Sequence Determination of Venom Components
2.4. Phylogenetic Affinities of the Calcins, Scorpines, La1-Like Peptides and Potassium Channel κ Toxins Found in the Transcriptomic Analysis of the Venom Gland of S. donensis
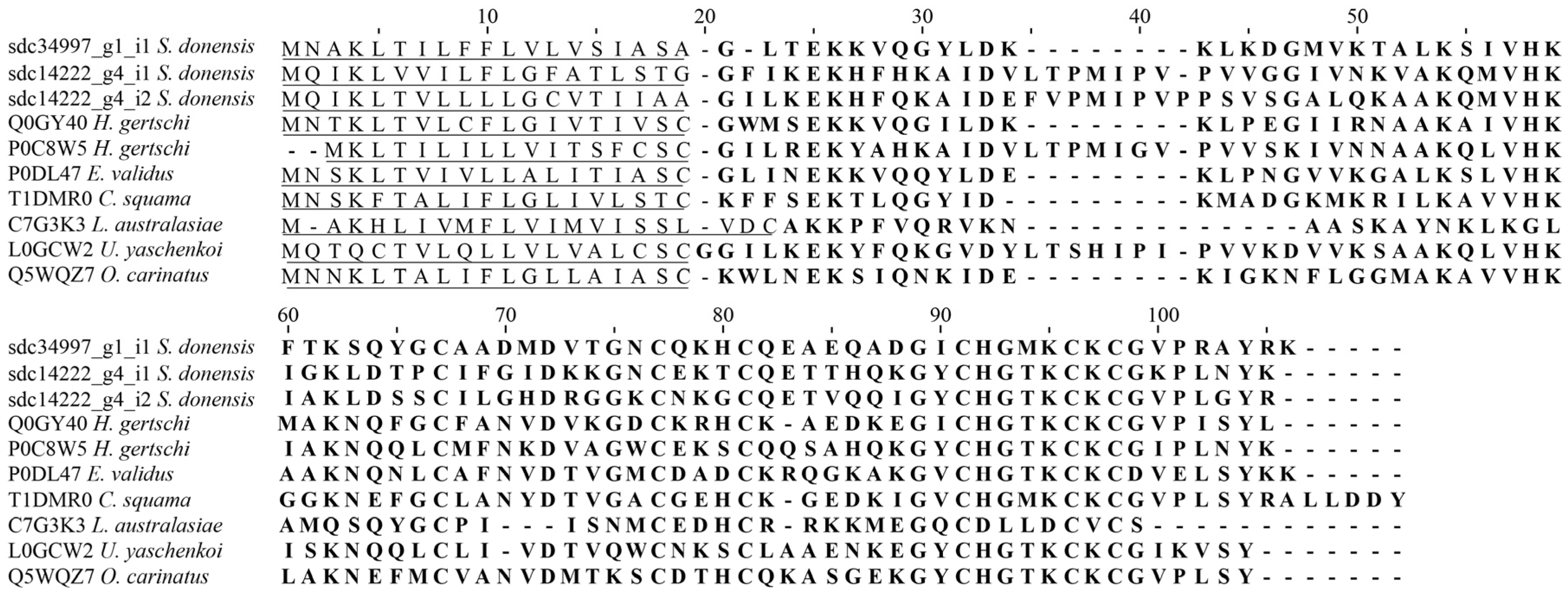
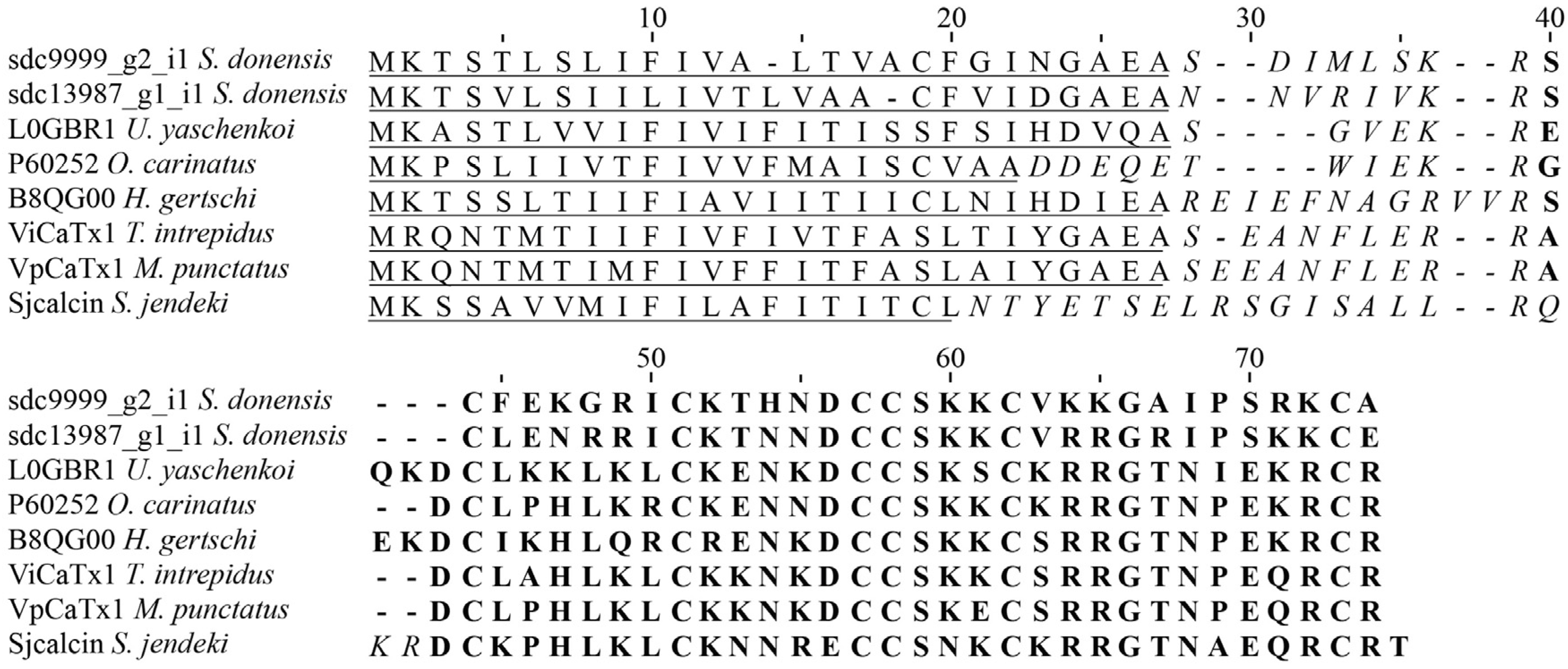
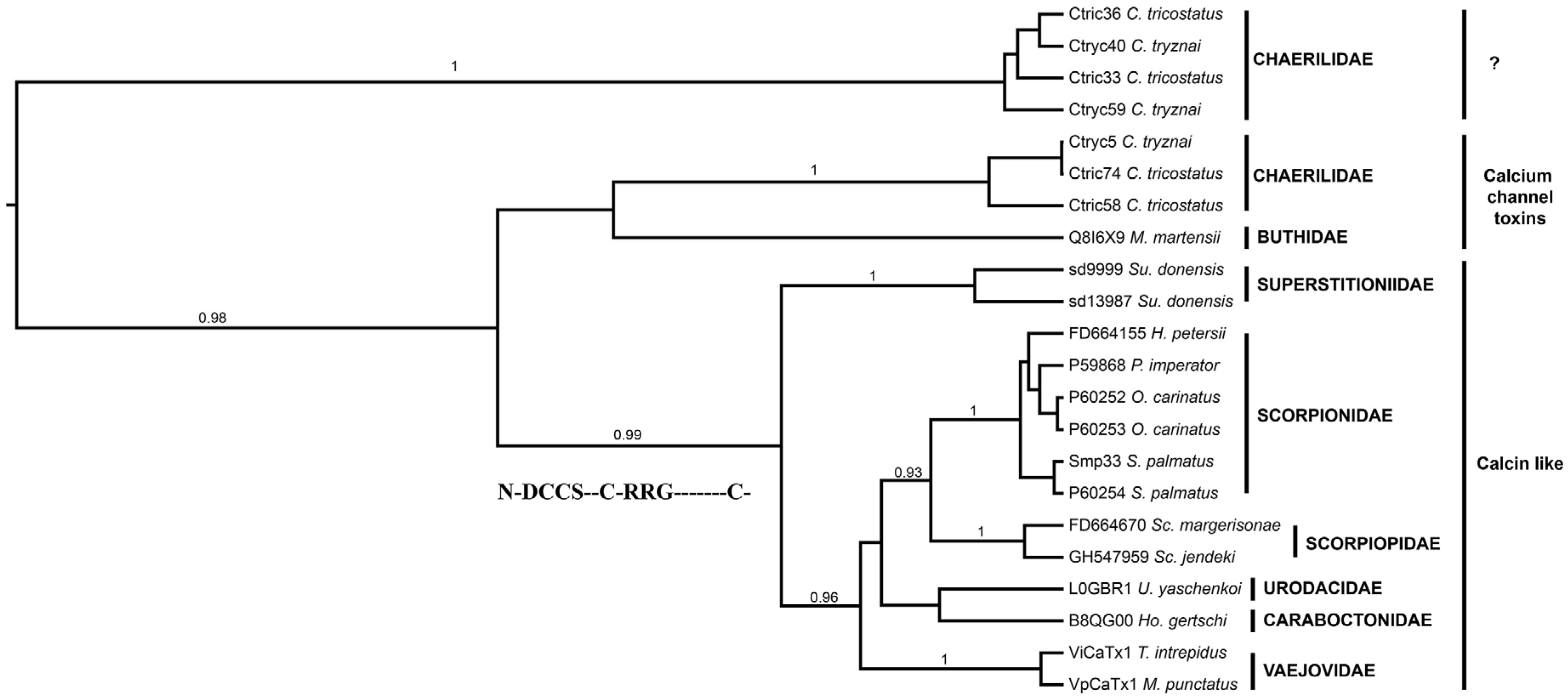
2.4.1. Calcins
2.4.2. Scorpines
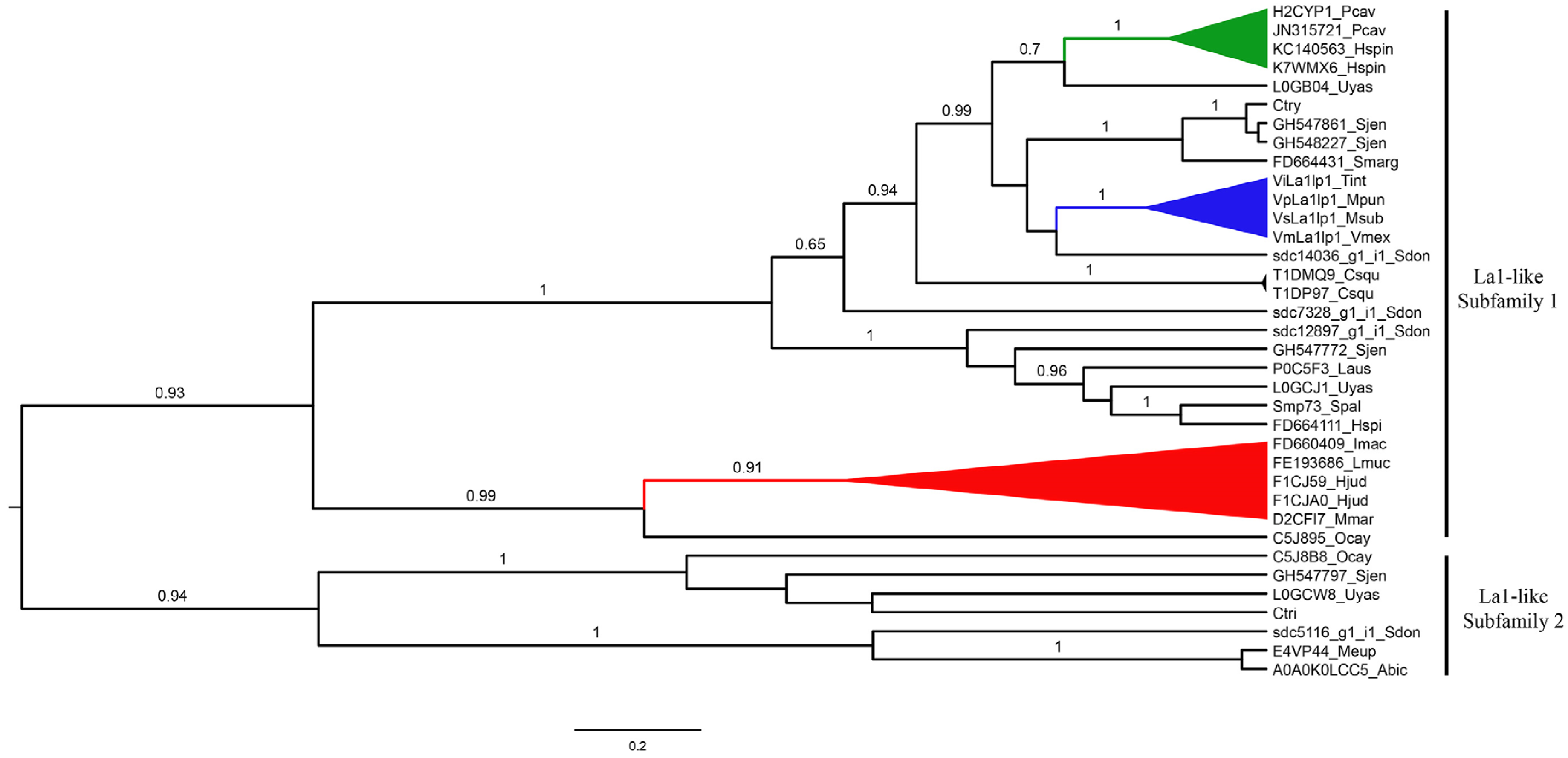
2.4.3. La1-Like Peptides
2.4.4. Potassium Channel κ Toxins (κKTx)
3. Conclusions
4. Materials and Methods
4.1. Biological Material
4.2. Molecular Mass Determination and Protein Identification
4.3. RNA Extraction, RNA-Seq and Venom Gland Transcriptome Assembly
4.4. Multiple Sequence Alignments, Phylogenetic Analysis and Motif Search
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Santibáñez-López, C.E.; Francke, O.F.; Ureta, C.; Possani, L.D. Scorpions from Mexico: From species diversity to venom complexity. Toxins 2016, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.P.; Férnandez, R.; Esposito, L.A.; Gónzalez-Santillán, E.; Monod, L. Phylogenomic resolution of scorpions reveals multivel discordance with morphological phylogenetic signal. Proc. R. Soc. B 2015, 282, 20142953. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; He, Y.; Zhao, R.; Wu, Y.; Li, W.; Cao, Z. Extreme diversity of scorpion venom peptides and proteins revealed by transcriptomic analysis: Implication for proteome evolution of scorpion venom arsenal. J. Proteom. 2012, 75, 1563–1576. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Undheim, E.A.; Chan, A.H.; Koludarov, I.; Muñoz-Gómez, S.A.; Antunes, A.; Fry, B.G. Evolution stings: The origin and diversification of scorpion toxin Peptide scaffolds. Toxins 2013, 5, 2456–2487. [Google Scholar] [CrossRef] [PubMed]
- Santibáñez-López, C.E.; Possani, L.D. Overview of the Knottin scorpion toxin-like peptides in scorpion venoms: Insights on their classification and evolution. Toxicon 2015, 107, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Luna-Ramírez, K.; Quintero-Hernández, V.; Juárez-González, V.R.; Possani, L.D. Whole transcriptome of the venom gland from Urodacus yaschenkoi scorpion. PLoS ONE 2015, 10, e0127883. [Google Scholar] [CrossRef] [PubMed]
- Luna-Ramírez, K.; Quintero-Hernández, V.; Vargas-Jaimes, L.; Batista, C.V.; Winkel, K.D.; Possani, L.D. Characterization of the venom from the Australian scorpion Urodacus yaschenkoi: Molecular mass analysis of components, cDNA sequences and peptides with antimicrobial activity. Toxicon 2013, 63, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Vignoli, V.; Prendini, L. Systematic revision of the trolgomorphic North American Scorpion family Typhlochactidae (Scorpiones: Chactoidae). Bull. Am. Mus. Nat. Hist. 2009, 326, 1–94. [Google Scholar] [CrossRef]
- Williams, S.C. Scorpions of Baja California, Mexico, and Adjacent Islands. Occ. Pap. Calif. Acad. Sci. 1980, 135, 1–127. [Google Scholar]
- Darbon, H. Animal toxin and ion channels. J. Soc. Biol. 1999, 193, 445–450. [Google Scholar] [PubMed]
- Quintero-Hernández, V.; Ramírez-Carreto, S.; Romero-Gutiérrez, M.T.; Valdez-Velázquez, L.L.; Becerril, B.; Possani, L.D.; Ortiz, E. Transcriptome analysis of scorpion species belonging to the Vaejovis genus. PLoS ONE 2015, 10, e0117188. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Gurrola, G.B.; Zhang, J.; Valdivia, C.R.; SanMartín, M.; Zamudio, F.Z.; Zhang, L.; Possani, L.D.; Valdivia, H.H. Structure-function relationships of peptides forming the calcin family of ryanodine receptor ligands. J. Gen. Phys. 2016, 147, 375–394. [Google Scholar] [CrossRef] [PubMed]
- Valdivia, H.H.; Fuentes, O.; El-Hayek, R.; Morrissette, J.; Coronado, R. Activation of the ryanodine receptor Ca2þ-release channel of sarcoplasmic reticulum by a novel scorpion venom. J. Biol. Chem. 1991, 266, 19135–19138. [Google Scholar] [PubMed]
- Morrissette, J.; Beurg, M.; Sukhareva, M.; Coronado, R. Purification and characterization of ryanotoxin, a peptide with actions similar to those of ryanodine. Biophys. J. 1996, 71, 707–721. [Google Scholar] [CrossRef]
- Ji, Y.H.; Liu, Y.; Yu, K.; Ohishi, T.; Hoshino, M.; Mochizuki, T.; Yanaihara, N. Amino acid sequence of an novel activator of ryanodine receptor on skeletal muscle. Chin. Sci. Bull. 1997, 42, 952–956. [Google Scholar] [CrossRef]
- Valdivia, H.H.; Kirby, M.S.; Lederer, W.J.; Coronado, R. Scorpion toxins targeted against the sarcoplasmic reticulum Ca2+- release channel of skeletal and cardiac muscle. Proc. Natl. Acad. Sci. USA 1992, 89, 12185–12189. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Hernández, V.; Jiménez-Vargas, J.M.; Gurrola, G.B.; Valdivia, H.H.; Possani, L.D. Scorpion venom components that affect ion-channels function. Toxicon 2013, 76, 328–342. [Google Scholar] [CrossRef] [PubMed]
- Conde, R.; Zamudio, F.Z.; Rodríguez, M.H.; Possani, L.D. Scorpine, an anti-malaria and anti-bacterial agent purified from scorpion venom. FEBS Lett. 2000, 471, 165–168. [Google Scholar] [CrossRef]
- Diego-García, E.; Schwartz, E.F.; D’Suze, G.; González, S.A.; Batista, C.V.; Garcia, B.I.; Rodríguez de la Vega, R.C.; Possani, L.D. Wide phylogenetic distribution of scorpine and long-chain β-KTx-like peptides in scorpion venoms: Identification of “orphan” components. Peptides 2007, 28, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Laraba-Djebari, F.; Adi-Bessalem, S.; Hammoudi-Triki, D. Scorpion Venoms: Pathogenesis and Biotherapies. In Scorpion Venoms; Gopalakrishnakone, P., Possani, L.D., Schwartz, E.F., Rodríguez de la Vega, R.C., Eds.; Springer: Dordrecht, The Netherlands, 2015; pp. 63–86. [Google Scholar]
- Carbone, E.; Wanke, E.; Prestipino, G.; Possani, L.D.; Maelicke, A. Selective blockage of voltage-dependent K+ channels by a novel scorpion toxin. Nature 1982, 296, 90–91. [Google Scholar] [CrossRef] [PubMed]
- Doyle, D.A.; Morais-Cabral, J.; Pfuetzner, R.A.; Kuo, A.; Gulbis, J.M.; Cohen, S.L.; Chait, B.R.; MacKinnon, R. The structure of potassium channel: Molecular basis of K+ conduction and selectivity. Science 1998, 280, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, E.F.; Diego-García, E.; Rodríguez de la Vega, R.C.; Possani, L.D. Transcriptome analysis of the venom gland of the Mexican scorpion Hadrurus gertschi (Arachnida: Scorpiones). BMC Genom. 2007, 8, 119. [Google Scholar] [CrossRef] [PubMed]
- Possani, L.D.; Becerril, B.; Delepierre, M.; Tytgat, J. Scorpion toxins specific for Na+-channels. Eur. J. Biochem. 1999, 264, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez de la Vega, R.C.; Possani, L.D. Overview of scorpion toxins specific for Na+ channels and related peptides: Biodiversity, structure-function relationships and evolution. Toxicon 2005, 46, 831–844. [Google Scholar] [CrossRef] [PubMed]
- InterPro: Scorpion Long Chain Toxin/Defensin (IPR002061). Available online: http://www.ebi.ac.uk/interpro/entry/IPR002061 (accessed on 17 March 2016).
- Guerrero-Vargas, J.A.; Mourao, C.B.; Quintero-Hernández, V.; Possani, L.D.; Schwartz, E.F. Identification and phylogenetic analysis of Tityus pachyurus and Tityus obscurus novel putative Na+-channel scorpion toxins. PLoS ONE 2012, 7, e30478. [Google Scholar] [CrossRef] [PubMed]
- Inceoglu, A.B.; Lango, J.; Pessah, I.N.; Hammock, B.D. Three structurally related, highly potent, peptides from the venom of Parabuthus transvaalicus possess divergent biological activity. Toxicon 2005, 45, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Inceoglu, A.B.; Lango, J.; Wu, J.; Hawkins, P.; Southern, J.; Hammock, B.D. Isolation and characterization of a novel type of neurotoxic peptide from the venom of the South African scorpion Parabuthus transvaalicus. Eur. J. Biochem. 2001, 268, 5407–5413. [Google Scholar] [CrossRef] [PubMed]
- Batista, C.V.F.; D’Suze, G.; Gómez-Lagunas, F.; Zamudio, F.Z.; Encarnacion, S.; Sevcik, C.; Possani, L.D. Proteomic analysis of Tityus discrepans scorpion venom and amino acid sequence of novel toxins. Proteomics 2006, 6, 3718–3727. [Google Scholar] [CrossRef] [PubMed]
- D’Suze, G.; Schwartz, E.F.; García-Gómez, B.I.; Sevcik, C.; Possani, L.D. Molecular cloning and nucleotide sequence analysis of genes from a cDNA library of the scorpion Tityus discrepans. Bichimie 2009, 91, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Ma, Y.; He, Y.; Di, Z.; Wu, Y.L.; Cao, Z.J.; Li, W.X. Comparative venom gland transcriptome analysis of the scorpion Lychas mucronatus reveals intraspecific toxic gene diversity and new venomous components. BMC Genom. 2010, 11, 452. [Google Scholar]
- García-Gómez, B.I.; Olamendi-Portugal, T.C.; Paniagua, J.; van der Walt, J.; Dyason, K.; Possani, L.D. Heterologous expression of a gene that codes for Pg8, a scorpion toxin of Parabuthus granulatus, capable of generating protecting antibodies in mice. Toxicon 2009, 53, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Soudani, N.; Gharbi-Chili, J.; Srairi-Abid, N.; Martin-El, Y.C.; Planells, R.; Margotat, A.; Torresani, J.; El Ayeb, M. Isolation and molecular characterization of LVP1 lipolysis activating peptide from scorpion Buthus occitanus tunetanus. Biochim. Biophys. Acta 2005, 1747, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez de la Vega, R.C.; Possani, L.D. Current views on scorpion toxins specific for K+-channels. Toxicon 2004, 43, 865–875. [Google Scholar] [CrossRef] [PubMed]
- InterPro: Scorpion Short Chain Toxin, Potassium Channel Inhibitor (IPR001947). Available online: http://www.ebi.ac.uk/interpro/entry/IPR002061 (accessed on 17 March 2016).
- InterPro: Long Chain Scorpion Toxin Family (IPR029237). Available online: http://www.ebi.ac.uk/interpro/entry/IPR029237 (accessed on 17 March 2016).
- Zhu, S.Y.; Huys, I.; Dyason, K.; Verdonck, F.; Tytgat, J. Evolutionary trace analysis of scorpion toxins specific for K-channels. Proteins 2004, 54, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Yin, S.; Ma, Y.; Wu, Y.L.; Zhao, R.; Gan, G.; Ding, J.; Cao, Z.; Li, W. Molecular cloning and electrophysiological studies on the first K(+) channel toxin (LmKTx8) derived from scorpion Lychas mucronatus. Peptides 2007, 28, 2306–2312. [Google Scholar] [CrossRef] [PubMed]
- Luna-Ramírez, K.; Bartok, A.; Restano-Cassulin, R.; Quintero-Hernández, V.; Coronas, F.; Christinseen, J.; Wright, C.E.; Panyi, G.; Possani, L.D. Structure, molecular modeling and function of Urotoxin, the first potassium channel blocker from the venom of Asutralian scorpion Urodacus yaschenkoi. Mol. Pharmacol. 2014, 86, 28–41. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, U.C.; Candido, D.M.; Coronado-Dorce, V.A.; Junquiera-de-Azevedo, I.D.L. The transcriptome recipe for the venom cocktail of Tityus bahiensis scorpion. Toxicon 2015, 95, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Vandendriessche, T.; Kopljar, I.; Jenkins, D.P.; Diego-García, E.; Abdel-Mottaleb, Y.; Vermassen, E.; Clynen, E.; Schoofs, L.; Wulff, H.; Snyders, D.; et al. Purification, molecular cloning and functional characterization of HelaTx1 (Heterometrus laoticus): The first member of a new kappa KTx. Biochem. Pharmacol. 2012, 83, 1307–1317. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, L.; Singh, J.; Humblet, C.; Guruprasad, K.; Blundell, T. Snail and spider toxins share a similar tertiary structure and ‘cystine motif’. Nat. Struct. Biol. 1994, 1, 850–852. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.Y.; Darbon, H.; Dyason, K.; Verdonck, F.; Tytgat, J. Evolutionary origin of inhibitor cysteine knot peptides. FASEB J. 2003, 17, 1765–1767. [Google Scholar] [PubMed]
- Kozlov, S.; Malyavka, A.; McCutchen, B.; Lu, A.; Schepers, E.; Hermann, R.; Grishin, E. A novel strategy for the identification of toxinlike structures in spider venom. Proteins 2005, 59, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Almaaytah, A.; Albalas, O. Scorpion venom peptides with non disulfide bridges: A review. Peptides 2014, 51, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, E.; Gurrola, G.B.; Schwartz, E.F.; Possani, L.D. Scorpion venom components as potential candidates for drug development. Toxicon 2015, 93, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Sherman, P.; Luo, L.; Bowie, J.; Zhu, S. Structural and functional characterization of two genetically related meucin peptides hihglights evolutionary divergence and convergence in antimicrobial peptides. FASEB J. 2009, 23, 1230–1245. [Google Scholar] [CrossRef] [PubMed]
- Farajzadeh-Sheikh, A.; Jolodar, A.; Ghaemmaghami, S. Sequence characterization of cDNA sequence of encoding of an antimicrobial peptide with no disulfide bridge from the Iranian Mesobuthus eupeus venomous gland. Red. Crescent. Med. J. 2013, 15, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Carreto, S.; Quintero-Hernández, V.; Jiménez-Vargas, J.M.; Corzo, G.; Possani, L.D.; Ortiz, E. Gene cloning and functional characterization of four novel antimicrobial-like peptides from scorpions of the family Vaejovidae. Peptides 2012, 34, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Li, Z.; Zhang, R.; Wu, Y.; Li, W.; Cao, Z. StCT2, a new antibacterial peptide characterized from the venom of the scorpion Scorpiops tibetanus. Peptides 2012, 36, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Carreto, S.; Jiménez-Vargas, J.M.; Rivas-Santiago, B.; Corzo, G.; Possani, L.D.; Becerril, B.; Ortiz, E. Peptides from the scorpion Vaejovis punctatus with broad antimicrobial activity. Peptides 2015, 73, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, M.; Otsuki, J.; Hanai, Y.; Nakagawa, Y.; Miyagawa, H. Characterization of peptide components in the venom of the scorpion Liocheles australasiae (Hemiscorpiidae). Toxicon 2007, 50, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.C.; Nie, Y.; Luo, X.; Wu, S.; Shi, W.; Zhang, L.; Liu, Y.; Cao, H.; Yang, Y.; Zhou, J. Molecular and bioinformatical characterization of a novel superfamily of cysteine-rich peptides from arthropods. Peptides 2013, 41, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.C.; Camargos, T.S.; Maranhao, A.Q.; Silva-Pereira, I.; Silva, L.P.; Possani, L.D.; Schwartz, E.F. Cloning and characterization of cDNA sequences encoding for new venom peptides of the Brazilian scorpion Opisthacanthus cayaporum. Toxicon 2009, 54, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhao, R.; He, Y.; Li, S.; Liu, J.; Wu, Y.; Cao, Z.; Li, W. Transcriptome analysis of the venom gland of the scorpion Scorpiops jendeki: Implications for the evolution of the scorpion venom arsenal. BMC Genom. 2009, 10, 290. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Gao, R.; Gopalakrishnakone, P. Isolation and characterization of a hyaluronidase from the venom of Chinese red scorpion Buthus martensi. Comp. Biochem. Physiol. 2008, 148C, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Horta, C.C.; Magalhaes, B.F.; Oliveira-Mendes, B.B.; do Carmo, A.O.; Duarte, C.G.; Felicori, L.F.; Machado-de-Avila, R.A.; Chavez-Olortegui, C.; Kalapothakis, E. Molecular, immunological, and biological characterization of Tityus serrulatus venom hyaluronidase: New insights into its role in envenomation. PLoS Negl. Trop. Dis. 2014, 8, e2693. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, B.; Yang, W.; Cao, Z.; Zhuo, R.; Li, W.; Wu, Y. SjAPI, the first functionally characterized Ascaris-Type protease inhibitor from animal venoms. PLoS ONE 2013, 8, e57529. [Google Scholar] [CrossRef] [PubMed]
- Gronenborn, A.M.; Nilges, M.; Peanasky, R.J.; Clore, G.M. Sequential resonance assignment and secondary structure determination of the Ascaris trypsin inhibitor, a member of a novel class of proteinase inhibitors. Biochemistry 1990, 29, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Diego-Garcia, E.; Peigneur, S.; Clynen, E.; Marien, T.; Czech, L.; Schoofs, L. Molecular diversity of the telson and venom components from Pandinus cavimanus (Scorpionidae Latreille 1802): Transcriptome, venomics and function. Proteomics 2012, 12, 313–328. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; Hu, Y.T.; Yang, W.S.; He, Y.W.; Feng, J.; Wang, B.; Zhao, R.M.; Ding, J.P.; Li, W.X.; Wu, Y.L. Hg1, novel peptide inhibitor specific for Kv1.3 channels from first scorpion kunitz.type potassium channel toxin family. J. Biol. Chem. 2012, 287, 13813–13821. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Deng, M.; Duan, Z.; Tang, X.; Liang, S. Molecular cloning, bioinformatics analysis and functional characterization of HWTX-XI toxin superfamily from the spider Ornithoctonus huwena. Peptides 2014, 59, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, G.M.; Roelants, K.; O’Bryan, M.K. The CAP superfamily: Cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins—roles in reproduction, cancer, and immune defense. Endocr. Rev. 2008, 29, 865–897. [Google Scholar] [CrossRef] [PubMed]
- Morgenstern, D.; Rohde, B.H.; King, G.F.; Tal, T.; Sher, D.; Zlotkin, E. The tale of a resting gland: Transcriptome of a replete venom gland from the scorpion Hottentotta judaicus. Toxicon 2011, 57, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shi, W.; Zeng, X.C.; Ge, F.; Yang, M.; Nie, Y.; Bao, A.; Wu, S. Unique diversity of the venom peptides from the scorpion Androctonus bicolor revealed by transcriptomic and proteomic analysis. J. Proteomics 2015, 128, 231–250. [Google Scholar] [CrossRef] [PubMed]
- Kotsyfakis, M.; Schwarz, A.; Erhart, J.; Ribeiro, J.M. Tissue- and time-dependent transcription in Ixodes ricinus salivary glands and midguts when blood feeding on the vertebrate host. Sci. Rep. 2015, 5, 9103. [Google Scholar] [CrossRef] [PubMed]
- Saucedo, A.L.; Flóres-Solis, D.; Rodríguez de la Vega, R.C.; Ramírez-Cordero, B.; Hernández-López, R.; Cano-Sánchez, P.; Noriega-Navarro, R.; García-Valdés, J.; Coronas-Valderrama, F.; de Roodt, A.; et al. New tricks of an old pattern: Structural versality of scorpion toxins with common cysteine spacing. J. Biol. Chem. 2012, 287, 12321–12330. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Mottaleb, Y.; Coronas, F.V.; de Roodt, A.R.; Possani, L.D.; Tytgat, J. A novel toxin from the venom of the scorpion Tityus trivittatus, is the first member of a new α-KTX subfamily. FEBS Lett. 2006, 582, 592–596. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotech. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alingment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 2005, 21, 2104–2105. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. Prottest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with beauti and the beast 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]


| Family | Venom Studies Available | cDNA or Transcriptome Analysis Available |
|---|---|---|
| Akravidae | No | No |
| Bothriuridae | No | Yes |
| Buthidae | Yes | Yes |
| Caraboctonidae | Yes | Yes |
| Chactidae | No | No |
| Chaerilidae | Yes | Yes |
| Diplocentridae | No | No |
| Euscorpiidae | Under revision * | Under revision * |
| Hemiscorpiidae | Yes | No |
| Heteroscorpionidae | No | No |
| Hormuridae | Yes | Yes |
| Iuridae | No | No |
| Pseudochactidae | No | No |
| Scorpionidae | Yes | Yes |
| Scorpiopidae | Yes | Yes |
| Superstitioniidae | This study | This study |
| Troglotayoscidae | No | No |
| Typhlochactidae | No | No |
| Urodacidae | Yes | Yes |
| Vaejovidae | Yes | Yes |
| Family | Calcins | Scorpines | La1-Like Peptides | Potassium Channel κ Toxins |
|---|---|---|---|---|
| Bothriuridae | No | Yes | Yes | No |
| Buthidae | Yes | Yes | Yes | No |
| Caraboctonidae | Yes | Yes | Yes | No |
| Chaerilidae | Yes | Yes | Yes | No |
| Hemiscorpiidae | No | No | No | No |
| Hormuridae | No | Yes | Yes | Yes |
| Scorpionidae | Yes | Yes | Yes | Yes |
| Scorpiopidae | Yes | Yes | Yes | No |
| Superstitioniidae | This study | This study | This study | This study |
| Urodacidae | Yes | Yes | Yes | No |
| Vaejovidae | Yes | Yes | Yes | Yes |
| Peptide Type | Transcript | Score | # Unique Pep. | Seq. Cov. | MW (kDa) | Protein/Accession |
|---|---|---|---|---|---|---|
| NaTx | sdc14462_g1_i1 | 257.51 | 7 | 91.43% | 11.7 | Lipolysis activating peptide 1 alpha chain/93140443 |
| sdc14462_g2_i2 | 29.7 | 2 | 29.81% | 11.8 | Birtoxin/20137305 | |
| sdc15193_g1_i1 | 20.23 | 1 | 85.71% | 7.1 | Toxin Cll7/31376362 | |
| sdc14462_g1_i2 | 118.64 | 1 | 78.10% | 11.8 | Altitoxin/116241245 | |
| KTx | sdc13949_g1_i1 | 27.98 | 2 | 65.38% | 5.5 | Toxin KTx 8/159146538 |
| sdc14273_g1_i2 | 58.09 | 1 | 76.32% | 4.1 | KTx 6.7/74838004 | |
| Sc | sdc14222_g4_i1 | 296.09 | 3 | 86.90% | 9.3 | Hg scorpine like 2/224493299 |
| sdc14222_g4_i2 | 153.55 | 3 | 76.47% | 9.2 | Hg scorpine like 2/224493299 | |
| NDBP | sdc12606_g1_i1 | 59.38 | 2 | 50.00% | 4 | Vejovine/325515699 |
| sdc14106_g1_i1 | 81.77 | 2 | 81.82% | 2.5 | Amp1/932534523 | |
| sdc28695_g1_i1 | 45.7 | 2 | 77.27% | 2.4 | Amp2/932534537 | |
| sdc4010_g1_i1 | 15.61 | 1 | 100.00% | 5.2 | Heterin 1/485896696 | |
| sdc13544_g1_i1 | 83.11 | 1 | 86.79% | 6.4 | CYLIP Cer 2/522802549 | |
| sdc14358_g5_i1 | 207.52 | 1 | 62.50% | 1.8 | Amphiphatic peptide CT2/384382524 | |
| sdc14358_g12_i1 | 211.79 | 1 | 52.38% | 2.3 | CYLIP Cer 2/522802549 | |
| sdc6540_g1_i1 | 161.85 | 1 | 52.38% | 2.3 | CYLIP Uro 3/522802596 | |
| La1-like | sdc13004_g1_i1 | 111.52 | 4 | 92.68% | 9.1 | Putative secreted protein/240247657 |
| sdc14036_g1_i1 | 162.81 | 2 | 100.00% | 8.4 | La1 like protein 15/430802826 | |
| sdc12897_g1_i1 | 50.14 | 2 | 74.32% | 7.9 | La1 like protein 13/430802824 | |
| sdc14589_g1_i1 | 53.02 | 1 | 90.24% | 8.8 | Toxin like protein 14/430802832 | |
| Enzymes | sdc14619_g1_i3 | 310.92 | 9 | 87.73% | 67.7 | Neprilysin 1/567441193 |
| sdc14393_g1_i1 | 70.24 | 4 | 76.13% | 26.4 | Phospholipase A2/218546750 | |
| sdc14212_g1_i1 | 90.57 | 3 | 74.19% | 24.1 | Phospholipase A2/218546750 | |
| sdc14619_g1_i1 | 42.67 | 1 | 78.17% | 83.3 | Neprilysin 1/567441193 | |
| CAP | sdc3852_g1_i1 | 97.67 | 3 | 71.20% | 43.1 | CAP-Uro-1/522802590 |
| sdc13900_g1_i1 | 87.21 | 2 | 54.10% | 38 | CAP-Iso-1/522802633 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santibáñez-López, C.E.; Cid-Uribe, J.I.; Batista, C.V.F.; Ortiz, E.; Possani, L.D. Venom Gland Transcriptomic and Proteomic Analyses of the Enigmatic Scorpion Superstitionia donensis (Scorpiones: Superstitioniidae), with Insights on the Evolution of Its Venom Components. Toxins 2016, 8, 367. https://doi.org/10.3390/toxins8120367
Santibáñez-López CE, Cid-Uribe JI, Batista CVF, Ortiz E, Possani LD. Venom Gland Transcriptomic and Proteomic Analyses of the Enigmatic Scorpion Superstitionia donensis (Scorpiones: Superstitioniidae), with Insights on the Evolution of Its Venom Components. Toxins. 2016; 8(12):367. https://doi.org/10.3390/toxins8120367
Chicago/Turabian StyleSantibáñez-López, Carlos E., Jimena I. Cid-Uribe, Cesar V. F. Batista, Ernesto Ortiz, and Lourival D. Possani. 2016. "Venom Gland Transcriptomic and Proteomic Analyses of the Enigmatic Scorpion Superstitionia donensis (Scorpiones: Superstitioniidae), with Insights on the Evolution of Its Venom Components" Toxins 8, no. 12: 367. https://doi.org/10.3390/toxins8120367








