Venomic Analysis of the Poorly Studied Desert Coral Snake, Micrurus tschudii tschudii, Supports the 3FTx/PLA2 Dichotomy across Micrurus Venoms
Abstract
:1. Introduction
2. Results and Discussion
2.1. The Venom Proteome of the Desert Coral Snake
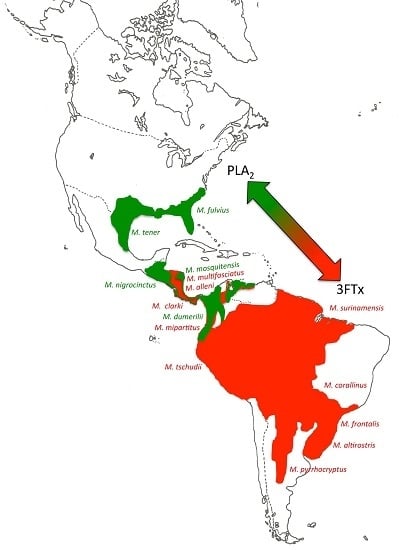
2.2. 3FTx- and PLA2-Predominant Venom Proteomes among Coral Snakes
2.3. Tracing the Evolutionary Origin of the 3FTx/PLA2 Dichotomy across Elapidae
3. Concluding Remarks and Perspectives
4. Materials and Methods
4.1. Venom
4.2. Isolation and Characterization of Venom Proteins
4.3. Characterization of the Venom Peptidome and Proteome
4.4. Determination of LD50 for Mice
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wagler, J. Serpentum Brasiliensium species novae, ou histoire naturelle des espèces nouvelles de serpens. In Animalia Nova Sive Species Novae; de Spix, J., Ed.; Franc. Seraph. Hübschmanni: Munich, Germany, 1824. [Google Scholar]
- Roze, J.A. Coral Snakes of the Americas. Biology, Identification and Venoms; Krieger Publishing Company: Malabar, FL, USA, 1996; p. 328. [Google Scholar]
- Campbell, J.A.; Lamar, W.W. The Venomous Reptiles of the Western Hemisphere; Cornell University Press: Ithaca, NY, USA, 2004; p. 476. [Google Scholar]
- The Reptile Database. Available online: http://www.reptile-database.org (accessed on 12 February2016).
- Pires, M.G.; da Silva, N.J., Jr.; Feitosa, D.T.; Prudente, A.L.; Filho, G.A.; Zaher, H. A new species of triadal coral snake of the genus Micrurus Wagler, 1824 (Serpentes: Elapidae) from northeastern Brazil. Zootaxa 2014, 3811, 569–584. [Google Scholar] [CrossRef] [PubMed]
- Feitosa, D.T.; da Silva, N.J.; Pires, M.G.; Zaher, H.; Prudente, A.L. A new species of monadal coral snake of the genus Micrurus (Serpentes, Elapidae) from western Amazon. Zootaxa 2015, 3974, 538–554. [Google Scholar] [CrossRef] [PubMed]
- Slowinski, J.B.; Keogh, J.S. Phylogenetic relationship of coral snakes based on cytochrome b mtDNA sequences. Mol. Phylogenet. Evol. 2005, 15, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Harvey, M.B.; Gutberlet, R.L. The evolution of New World venomous snakes. In The Venomous Reptiles of the Western Hemisphere; Campbell, J.A., Lamar, W.W., Eds.; Cornell University Press: Ithaca, NY, USA, 2004; pp. 634–682. [Google Scholar]
- Campbell, J.A.; Lamar, W.W. The Venomous Reptiles of Latin America; Cornell University Press: Ithaca, NY, USA; London, UK, 1989; p. 425. [Google Scholar]
- Savage, J.M.; Slowinski, J.B. The coloration of the venomous coral snakes (family Elapidae) and their mimics (families Aniliidae and Colubridae). Biol. J. Linn. Soc. 1992, 45, 235–254. [Google Scholar] [CrossRef]
- Slowinski, J.B. A phylogenetic analysis of the New World coral snakes (Elapidae: Leptomicrurus, Micruroides and Micrurus) based on allozymic and morphological characters. J. Herpetol. 1995, 29, 325–338. [Google Scholar] [CrossRef]
- Silva, N.J., Jr.; Sites, J.W., Jr. Phylogeny of South American triad coral snakes (Elapidae: Micrurus) based on molecular characters. Herpetologica 2001, 57, 1–22. [Google Scholar]
- Calvete, J.J. Snake venomics: From the inventory of toxins to biology. Toxicon 2013, 75, 44–62. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J. Next-generation snake venomics: Protein-locus resolution through venom proteome decomplexation. Expert Rev. Proteom. 2014, 11, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Brahma, R.K.; McCleary, R.J.; Kini, R.M.; Doley, R. Venom gland transcriptomics for identifying, cataloging, and characterizing venom proteins in snakes. Toxicon 2015, 93, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Lomonte, B. A bright future for integrative venomics. Toxicon 2015, 107, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Morgenstern, D.; Reitzel, A.M.; Moran, Y. Ecological venomics: How genomics, transcriptomics and proteomics can shed new light on the ecology and evolution of venom. J. Proteom. 2016, 135, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.; Vargas-Vargas, N.; Pla, D.; Sasa, M.; Rey-Suárez, P.; Sanz, L.; Gutiérrez, J.M.; Calvete, J.J.; Lomonte, B. Snake venomics of Micrurus alleni and Micrurus mosquitensis from the Caribbean region of Costa Rica reveals two divergent compositional patterns in New World elapids. Toxicon 2015, 107, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Warrell, D.A. Snakebites in central and South America: Epidemiology, clinical features, clinical management. In The Venomous Reptiles of the Western Hemisphere; Campbell, J.A., Lamar, W.W., Eds.; Cornell University Press: Ithaca, NY, USA, 2004; pp. 709–761. [Google Scholar]
- Bucaretchi, F.; de Capitani, E.M.; Vieira, R.J.; Rodrigues, C.K.; Zannin, M.; da Silva, N.J., Jr.; Casais-E-Silva, L.L.; Hyslop, S. Coral snake bites (Micrurus spp.) in Brazil: A review of literature reports. Clin. Toxicol. 2016, 25, 1–13. [Google Scholar]
- Norris, R.L.; Pfalzgraf, R.R.; Laing, G. Death following coral snake bite in the United States—First documented case (with ELISA confirmation of envenomation) in over 40 years. Toxicon 2009, 53, 693–697. [Google Scholar] [CrossRef] [PubMed]
- Jan, G. Plan d’une iconographie descriptive des ophidiens et description sommaire de nouvelles espèces deserpents. Rev. Mag. Zool. Pure Apliquée (Paris) 1858, 2, 438–439 and 514–527. [Google Scholar]
- Silva, N.J., Jr.; Sites, J.W., Jr. Revision of the Micrurus frontalis complex (Serpentes: Elapidae). Herpetol. Monogr. 1999, 13, 142–194. [Google Scholar] [CrossRef]
- Peters, J.A.; Orejas-Miranda, B. Catalogue of the neotropical Squamata: Part I. Snakes. US Natl. Mus. Bull. 1970, 297, 1–347. [Google Scholar] [CrossRef]
- Mackessy, S.P.; Tu, A.T. Biology of the sea snakes and biochemistry of their venoms. In Toxin-Related Diseases: Poisons Originating from Plants, Animals and Spoilage; Tu, A.T., Ed.; Oxford IBH Publishing Co.: New Delhi, India, 1993; pp. 305–351. [Google Scholar]
- Jackson, T.N.; Sunagar, K.; Undheim, E.A.; Koludarov, I.; Chan, A.H.; Sanders, K.; Ali, S.A.; Hendrikx, I.; Dunstan, N.; Fry, B.G. Venom down under: Dynamic evolution of Australian elapid snake toxins. Toxins 2013, 5, 2621–2655. [Google Scholar] [CrossRef] [PubMed]
- Kini, R.M.; Doley, R. Structure, function and evolution of three-finger toxins: Mini proteins with multiple targets. Toxicon 2010, 56, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Utkin, Y.N. Three-finger toxins, a deadly weapon of elapid venom-milestones of discovery. Toxicon 2013, 62, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Du, X.-Y.; Clemetson, K.J. Snake venom l-amino acid oxidases. Toxicon 2002, 40, 659–665. [Google Scholar] [CrossRef]
- Filippovich, I.; Sorokina, N.; Masci, P.P.; de Jersey, J.; Whitaker, A.N.; Winzor, D.J.; Gaffney, P.J.; Lavin, M.F. A family of textilinin genes, two of which encode proteins with antihaemorrhagic properties. Br. J. Haematol. 2002, 119, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Bohlen, C.J.; Chesler, A.T.; Sharif-Naeini, R.; Medzihradszky, K.F.; Zhou, S.; King, D.; Sánchez, E.E.; Burlingame, A.L.; Basbaum, A.I.; Julius, D. A heteromeric Texas coral snake toxin targets acid-sensing ion channels to produce pain. Nature 2011, 479, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Baconguis, I.; Bohlen, C.J.; Goehring, A.; Julius, D.; Gouaux, E. X-ray structure of acid-sensing ion channel 1-snake toxin complex reveals open state of a Na(+)-selective channel. Cell 2014, 156, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Bohlen, C.J.; Julius, D. Receptor-targeting mechanisms of pain-causing toxins: How ow? Toxicon 2012, 60, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Aird, S.D. Ophidian envenomation strategies and the role of purines. Toxicon 2002, 40, 335–393. [Google Scholar] [CrossRef]
- Guo, C.; Liu, S.; Yao, Y.; Zhang, Q.; Sun, M.-Z. Past decade study of snake vemom l-amino acid oxidase. Toxicon 2012, 60, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.Y.; Sanders, K.L.; King, B.; Palci, A. Diversification rates and phenotypic evolution in venomous snakes (Elapidae). R. Soc. Open Sci. 2016, 3, 150277. [Google Scholar] [CrossRef] [PubMed]
- Greene, H.W. Dietary correlates of the origin and radiation of snakes. Am. Zool. 1983, 23, 431–441. [Google Scholar] [CrossRef]
- Daltry, J.C.; Wüster, W.; Thorpe, R.S. Diet and snake venom evolution. Nature 1996, 379, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Barlow, A.; Pook, C.E.; Harrison, R.A.; Wüster, W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. Biol. Sci. 2009, 276, 2443–2449. [Google Scholar] [CrossRef] [PubMed]
- Rey-Suárez, P.; Núñez, V.; Fernández, J.; Lomonte, B. Integrative characterization of the venom of the coral snake Micrurus dumerilii (Elapidae) from Colombia: Proteome, toxicity, and cross-neutralization by antivenom. J. Proteom. 2016, 136, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Corrêa-Netto, C.; Junqueira-de-Azevedo, I.L.; Silva, D.A.; Ho, P.L.; Leitão-de-Araújo, M.; Alves, M.L.; Sanz, L.; Foguel, D.; Zingali, R.B.; Calvete, J.J. Snake venomics and venom gland transcriptomic analysis of Brazilian coral snakes, Micrurus altirostris and M. corallinus. J. Proteom. 2011, 74, 1795–1809. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.; Alape-Girón, A.; Angulo, Y.; Sanz, L.; Gutiérrez, J.M.; Calvete, J.J.; Lomonte, B. Venomic and antivenomic analyses of the Central American coral snake, Micrurus nigrocinctus (Elapidae). J. Proteome Res. 2011, 10, 1816–1827. [Google Scholar] [CrossRef] [PubMed]
- Vergara, I.; Pedraza-Escalona, M.; Paniagua, D.; Restano-Cassulini, R.; Zamudio, F.; Batista, C.V.; Possani, L.D.; Alagón, A. Eastern coral snake Micrurus fulvius venom toxicity in mice is mainly determined by neurotoxic phospholipases A2. J. Proteom. 2014, 105, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Margres, M.J.; Aronow, K.; Loyacano, J.; Rokyta, D.R. The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genom. 2013, 14, 531. [Google Scholar] [CrossRef] [PubMed]
- Bénard-Valle, M.; Carbajal-Saucedo, A.; de Roodt, A.; López-Vera, E.; Alagón, A. Biochemical characterization of the venom of the coral snake Micrurus tener and comparative biological activities in the mouse and a reptile model. Toxicon 2014, 77, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Sasa, M.; Rey-Suárez, P.; Bryan, W.; Gutiérrez, J.M. Venom of the coral snake Micrurus clarki: Proteomic profile, toxicity, immunological cross-neutralization, and characterization of a three-finger toxin. Toxins 2016, 8, 138. [Google Scholar] [CrossRef] [PubMed]
- Castoe, T.A.; Smith, E.N.; Brown, R.M.; Parkinson, K.L. Higher-level phylogeny of Asian and American coralsnakes, their placement within the Elapidae (Squamata), and the systematic affinities at the enigmatic Asian coralsnake Hemibungarus calligaster (Weigmann, 1834). Zool. J. Linn. Soc. 2007, 151, 809–831. [Google Scholar] [CrossRef]
- Renjifo, C.; Smith, E.N.; Hodgson, W.C.; Renjifo, J.M.; Sanchez, A.; Acosta, R.; Maldonado, J.H.; Rivero, A. Neuromuscular activity of the venoms of the Colombian coral snakes Micrurus dissoleucus and Micrurus mipartitus: An evolutionary perspective. Toxicon 2012, 59, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Olamendi-Portugal, T.; Batista, C.; Restano-Cassulini, R.; Pando, V.; Villa-Hernandez, O.; Zavaleta-Martínez-Vargas, A.; Salas-Arruz, M.C.; Rodríguez de la Vega, R.C.; Becerril, B.; Possani, L. Proteomic analysis of the venom from the fish eating coral snake Micrurus surinamensis: Novel toxins, their function and phylogeny. Proteomics 2008, 8, 1919–1932. [Google Scholar] [CrossRef] [PubMed]
- Ciscotto, P.H.; Rates, B.; Silva, D.A.; Richardson, M.; Silva, L.P.; Andrade, H.; Donato, M.F.; Cotta, G.A.; Maria, W.S.; Rodrigues, R.J.; et al. Venomic analysis and evaluation of antivenom cross-reactivity of South American Micrurus species. J. Proteom. 2011, 74, 1810–1825. [Google Scholar] [CrossRef] [PubMed]
- Dokmetjian, J.C.; Del Canto, S.; Vinzón, S.; de Jiménez Bonino, M.B. Biochemical characterization of the Micrurus pyrrhocryptus venom. Toxicon 2009, 53, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Rey-Suárez, P.; Núñez, V.; Gutiérrez, J.M.; Lomonte, B. Proteomic and biological characterization of the venom of the redtail coral snake, Micrurus mipartitus (Elapidae), from Colombia and Costa Rica. J. Proteom. 2011, 75, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, G.D.; Furtado, M.F.; Portaro, F.C.; Sant’Anna, O.A.; Tambourgi, D.V. Diversity of Micrurus snake species related to their venom toxic effects and the prospective of antivenom neutralization. PLoS Negl. Trop. Dis. 2010, 4, e622. [Google Scholar] [CrossRef] [PubMed]
- Salazar, A.M.; Vivas, J.; Sánchez, E.E.; Rodríguez-Acosta, A.; Ibarra, C.; Gil, A.; Carvajal, Z.; Girón, M.E.; Estrella, A.; Navarrete, L.F.; et al. Hemostatic and toxinological diversities in venom of Micrurus tener tener, Micrurus fulvius fulvius and Micrurus isozonus coral snakes. Toxicon 2011, 58, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, I.; Sánchez, E.E.; Girón, M.E.; Estrella, A.; Guerrero, B.; Rodriguez-Acosta, F.A. Coral snake antivenom produced in chickens (Gallus domesticus). Rev. Inst. Med. Trop. São Paulo 2014, 56, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Gloss, A.D.; Whiteman, N.K. Balancing Selection: Walking a Tightrope. Curr. Biol. 2016, 26, R73–R76. [Google Scholar] [CrossRef] [PubMed]
- Sanders, K.L.; Lee, M.S.Y. Molecular evidence for a rapid late-Miocene radiation of Australasian venomous snakes. Mol. Phylogenet. Evol. 2008, 46, 1165–1173. [Google Scholar] [CrossRef] [PubMed]
- Pyron, R.A.; Burbrink, F.T.; Wiens, J.J. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 2013, 13, 93. [Google Scholar] [CrossRef] [PubMed]
- Sanders, K.L.; Lee, M.S.Y.; Mumpuni, B.T.; Rasmussen, A.R. Multilocus phylogeny and recent rapid radiation of the viviparous sea snakes (Elapidae: Hydrophiinae). Mol. Phylogenet. Evol. 2013, 66, 575–591. [Google Scholar] [CrossRef] [PubMed]
- Laustsen, A.H.; Gutiérrez, J.M.; Rasmussen, A.R.; Engmark, M.; Gravlund, P.; Sanders, K.L.; Lohse, B.; Lomonte, B. Danger in the reef: Proteome, toxicity, and neutralization of the venom of the olive sea snake, Aipysurus laevis. Toxicon 2015, 107, 187–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, C.H.; Tan, K.Y.; Lim, S.E.; Tan, N.H. Venomics of the beaked sea snake, Hydrophis schistosus: A minimalist toxin arsenal and its cross-neutralization by heterologous antivenoms. J. Proteom. 2015, 126, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Ghezellou, P.; Paiva, O.; Matainaho, T.; Ghassempour, A.; Goudarzi, H.; Kraus, F.; Sanz, L.; Williams, D.J. Snake venomics of two poorly known Hydrophiinae: Comparative proteomics of the venoms of terrestrial Toxicocalamus longissimus and marine Hydrophis cyanocinctus. J. Proteom. 2012, 75, 4091–4101. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Pla, D.; Sasa, M.; Tsai, W.C.; Solórzano, A.; Ureña-Díaz, J.M.; Fernández-Montes, M.L.; Mora-Obando, D.; Sanz, L.; Gutiérrez, J.M.; et al. Two color morphs of the pelagic yellow-bellied sea snake, Pelamis platura, from different locations of Costa Rica: Snake venomics, toxicity, and neutralization by antivenom. J. Proteom. 2014, 103, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Pahari, S.; Bickford, D.; Fry, B.G.; Kini, R.M. Expression pattern of three-finger toxin and phospholipase A2 genes in the venom glands of two sea snakes, Lapemis curtus and Acalyptophis peronii: Comparison of evolution of these toxins in land snakes, sea kraits and sea snakes. BMC Evol. Biol. 2007, 7, 175. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Fung, S.Y.; Yap, M.K.; Leong, P.K.; Liew, J.L.; Tan, N.H. Unveiling the elusive and exotic: Venomics of the Malayan blue coral snake (Calliophis bivirgata flaviceps). J. Proteom. 2016, 132, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mackessy, S.P. Venom composition in rattlesnakes: trends and biological significance. In The Biology of Rattlesnakes; Hayes, W.K., Beaman, K.R., Cardwell, M.D., Bush, S.P., Eds.; Loma Linda University Press: Loma Linda, CA, USA, 2008; pp. 495–510. [Google Scholar]
- Calvete, J.J.; Sanz, L.; Cid, P.; de la Torre, P.; Flores-Díaz, M.; Dos Santos, M.C.; Borges, A.; Bremo, A.; Angulo, Y.; Lomonte, B.; et al. Snake venomics of the Central American rattlesnake Crotalus simus and the South American Crotalus durissus complex points to neurotoxicity as an adaptive paedomorphic trend along Crotalus dispersal in South America. J. Proteome Res. 2010, 9, 528–544. [Google Scholar] [CrossRef] [PubMed]
- Durban, J.; Pérez, A.; Sanz, L.; Gómez, A.; Bonilla, F.; Rodríguez, S.; Chacón, D.; Sasa, M.; Angulo, Y.; Gutiérrez, J.M.; et al. Integrated “omics” profiling indicates that miRNAs are modulators of the ontogenetic venom composition shift in the Central American rattlesnake, Crotalus simus simus. BMC Genom. 2013, 14, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massey, D.J.; Calvete, J.J.; Sánchez, E.E.; Sanz, L.; Richards, K.; Curtis, R.; Boesen, K. Venom variability and envenoming severity outcomes of the Crotalus scutulatus scutulatus (Mojave rattlesnake) from Southern Arizona. J. Proteom. 2012, 75, 2576–2587. [Google Scholar] [CrossRef] [PubMed]
- Diz, A.P.; Martínez-Fernández, M.; Rolán-Alvarez, E. Proteomics in evolutionary ecology: Linking the genotype with the phenotype. Mol. Ecol. 2012, 21, 1060–1080. [Google Scholar] [CrossRef] [PubMed]
- Baer, B.; Millar, A.H. Proteomics in evolutionary ecology. J. Proteom. 2016, 135, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Diz, A.P.; Calvete, J.J. Ecological proteomics: Is the field ripe for integrating proteomics into evolutionary ecology research? J. Proteom. 2016, 135, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Eichberg, S.; Sanz, L.; Calvete, J.J.; Pla, D. Constructing comprehensive venom proteome reference maps for integrative venomics. Expert Rev. Proteom. 2015, 12, 557–573. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Juárez, P.; Sanz, L. Snake venomics, strategy and aplications. J. Mass Spectrom. 2007, 42, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Finney, D.J. Statistical Methods in Biological Assay; Charles Griffin and Company Ltd.: London, UK, 1971. [Google Scholar]


| 3FTx-Rich Venom | LD50 | Reference | PLA2-Rich Venom | LD50 | References |
|---|---|---|---|---|---|
| M. altirostris | i.p. 0.26–0.65 | [41] | M. fulvius | i.v. 0.32 ± 0.12 | [43] |
| i.p. 2.60–4.40 | [53] | ||||
| M. corallinus | i.p. 0.25–1.35 | [41] | M. tener | s.c. 4.4 | [45] |
| i.v. 0.78 ± 0.14 | [54,55] | ||||
| M. mipartitus | i.p. 0.47 | [52] | M. nigrocinctus | i.v. 0.3–0.5 | [45] |
| i.p. 0.4–1.2 | |||||
| s.c. 1.7–2.5 | |||||
| M. multifasciatus | i.p. 1.35 | [52] | M. mosquitensis | i.v. 0.20–0.61 | [18] |
| M. alleni | i.v. 0.23–0.55 | [18] | M. dumerilii | i.p. 0.8–1.9 | [40] |
| i.v. 0.74 ± 0.16 | |||||
| M. tschudii | i.p. 0.44–0.81 | [tw] | |||
| M. frontalis | i.p. 0.20–1.45 | [53] | |||
| M. pyrrhocryptus | i.p. 1.10 ± 0.10 | [51] | |||
| M. surinamensis | i.p. 2.15–4.35 | [53] | |||
| M. clarki | i.v. 0.42–1.38 | [46] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanz, L.; Pla, D.; Pérez, A.; Rodríguez, Y.; Zavaleta, A.; Salas, M.; Lomonte, B.; Calvete, J.J. Venomic Analysis of the Poorly Studied Desert Coral Snake, Micrurus tschudii tschudii, Supports the 3FTx/PLA2 Dichotomy across Micrurus Venoms. Toxins 2016, 8, 178. https://doi.org/10.3390/toxins8060178
Sanz L, Pla D, Pérez A, Rodríguez Y, Zavaleta A, Salas M, Lomonte B, Calvete JJ. Venomic Analysis of the Poorly Studied Desert Coral Snake, Micrurus tschudii tschudii, Supports the 3FTx/PLA2 Dichotomy across Micrurus Venoms. Toxins. 2016; 8(6):178. https://doi.org/10.3390/toxins8060178
Chicago/Turabian StyleSanz, Libia, Davinia Pla, Alicia Pérez, Yania Rodríguez, Alfonso Zavaleta, Maria Salas, Bruno Lomonte, and Juan J. Calvete. 2016. "Venomic Analysis of the Poorly Studied Desert Coral Snake, Micrurus tschudii tschudii, Supports the 3FTx/PLA2 Dichotomy across Micrurus Venoms" Toxins 8, no. 6: 178. https://doi.org/10.3390/toxins8060178
APA StyleSanz, L., Pla, D., Pérez, A., Rodríguez, Y., Zavaleta, A., Salas, M., Lomonte, B., & Calvete, J. J. (2016). Venomic Analysis of the Poorly Studied Desert Coral Snake, Micrurus tschudii tschudii, Supports the 3FTx/PLA2 Dichotomy across Micrurus Venoms. Toxins, 8(6), 178. https://doi.org/10.3390/toxins8060178