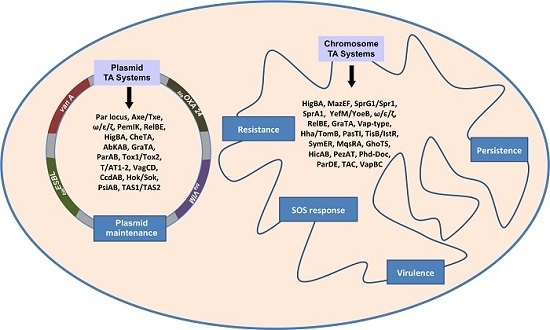
Toxin-Antitoxin Systems in Clinical Pathogens
Abstract
:1. Introduction
1.1. Pathogens in the ESKAPE Group
1.1.1. Enterococcus spp.
1.1.2. Staphylococcus aureus
1.1.3. Klebsiella pneumoniae
1.1.4. Acinetobacter baumannii
1.1.5. Pseudomonas aeruginosa
1.1.6. Enterobacter spp.
1.2. Other Pathogens
1.2.1. Escherichia coli
1.2.2. Burkholderia spp.
1.2.3. Streptococcus pneumoniae
1.2.4. Mycobacterium tuberculosis
2. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct. 2009, 4. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Park, J.H.; Inouye, M. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 2011, 45, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Mutschler, H.; Gebhardt, M.; Shoeman, R.L.; Meinhart, A. A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol. 2011, 9. [Google Scholar] [CrossRef] [PubMed]
- Unoson, C.; Wagner, E.G. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 2008, 70, 258–270. [Google Scholar] [CrossRef] [PubMed]
- Leplae, R.; Geeraerts, D.; Hallez, R.; Guglielmini, J.; Drèze, P.; Van Melderen, L. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011, 39, 5513–5525. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Rasmussen, P.B.; Molin, S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 1986, 83, 3116–3120. [Google Scholar] [CrossRef] [PubMed]
- Van Melderen, L.; Saavedra De Bast, M. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 2009, 5. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, A.; Yahara, K.; Kobayashi, I.; Iwasa, Y. Genetic addiction: selfish gene's strategy for symbiosis in the genome. Genetics. 2006, 172, 1309–1323. [Google Scholar] [CrossRef] [PubMed]
- Van Melderen, L. Toxin-antitoxin systems: why so many, what for? Curr. Opin. Microbiol. 2010, 13, 781–785. [Google Scholar] [CrossRef] [PubMed]
- Lehnherr, H.; Maguin, E.; Jafri, S.; Yarmolinsky, M.B. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 1993, 233, 414–428. [Google Scholar] [CrossRef] [PubMed]
- Ramage, H.R.; Connolly, L.E.; Cox, J.S. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009, 5. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Inouye, M. mRNA interferases, sequence-specific endoribonucleases from the toxin-antitoxin systems. Prog. Mol. Biol. Transl. Sci. 2009, 85, 467–500. [Google Scholar]
- Schuster, C.F.; Bertram, R. Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol. Lett. 2013, 340, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lord, D.M.; Cheng, H.Y.; Osbourne, D.O.; Hong, S.H.; Sanchez-Torres, V.; Quiroga, C.; Zheng, K.; Herrmann, T.; Peti, W.; et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 2012, 8, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Fair, R.J.; Tor, Y. Antibiotics and bacterial resistance in the 21st century. Perspect. Medicin. Chem. 2014, 6, 25–64. [Google Scholar] [PubMed]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Wang, X.; Ma, Q.; Zhang, X.S.; Wood, T.K. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J. Bacteriol. 2009, 191, 1258–1267. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Maisonneuve, E. Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 2012, 66, 103–123. [Google Scholar] [CrossRef] [PubMed]
- Kolodkin-Gal, I.; Verdiger, R.; Shlosberg-Fedida, A.; Engelberg-Kulka, H. A differential effect of E. coli toxin-antitoxin systems on cell death in liquid media and biofilm formation. PLoS ONE 2009, 4. [Google Scholar] [CrossRef] [PubMed]
- Brantl, S. Bacterial type I toxin-antitoxin systems. RNA Biol. 2012, 9, 1488–1490. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Weaver, K.E. Addiction toxin Fst has unique effects on chromosome segregation and cell division in Enterococcus faecalis and Bacillus subtilis. J. Bacteriol. 2006, 188, 5374–5384. [Google Scholar] [CrossRef] [PubMed]
- Halvorsen, E.M.; Williams, J.J.; Bhimani, A.J.; Billings, E.A.; Hergenrother, P.J. Txe, an endoribonuclease of the enterococcal Axe-Txe toxin-antitoxin system, cleaves mRNA and inhibits protein synthesis. Microbiology 2011, 157, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Sletvold, H.; Johnsen, P.J.; Hamre, I.; Simonsen, G.S.; Sundsfjord, A.; Nielsen, K.M. Complete sequence of Enterococcus faecium pVEF3 and the detection of an omega-epsilon-zeta toxin-antitoxin module and an ABC transporter. Plasmid 2008, 60, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Rosvoll, T.C.; Pedersen, T.; Sletvold, H.; Johnsen, P.J.; Sollid, J.E.; Simonsen, G.S.; Jensen, L.B.; Nielsen, K.M.; Sundsfjord, A. PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501- and pHTbeta-related replicons associated with glycopeptide resistance and stabilizing toxin-antitoxin systems. FEMS Immunol. Med. Microbiol. 2010, 58, 254–268. [Google Scholar] [CrossRef] [PubMed]
- Soheili, S.; Ghafourian, S.; Sekawi, Z.; Neela, V.K.; Sadeghifard, N.; Taherikalani, M.; Khosravi, A.; Ramli, R.; Hamat, R.A. The mazEF toxin-antitoxin system as an attractive target in clinical isolates of Enterococcus faecium and Enterococcus faecalis. Drug Des. Devel. Ther. 2015, 9, 2553–2561. [Google Scholar] [PubMed]
- Bukowski, M.; Lyzen, R.; Helbin, W.M.; Bonar, E.; Szalewska-Palasz, A.; Wegrzyn, G.; Dubin, G.; Dubin, A.; Wladyka, B. A regulatory role for Staphylococcus aureus toxin-antitoxin system PemIKSa. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Pinel-Marie, M.L.; Brielle, R.; Felden, B. Dual toxic-peptide-coding Staphylococcus aureus RNA under antisense regulation targets host cells and bacterial rivals unequally. Cell. Rep. 2014, 7, 424–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster, C.F.; Bertram, R. Toxin-Antitoxin Systems of Staphylococcus aureus. Toxins (Basel) 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Sayed, N.; Nonin-Lecomte, S.; Réty, S.; Felden, B. Functional and structural insights of a Staphylococcus aureus apoptotic-like membrane peptide from a toxin-antitoxin module. J. Biol. Chem. 2012, 287, 43454–43463. [Google Scholar] [CrossRef] [PubMed]
- Donegan, N.P.; Cheung, A.L. Regulation of the mazEF toxin-antitoxin module in Staphylococcus aureus and its impact on sigB expression. J. Bacteriol. 2009, 191, 2795–2805. [Google Scholar] [CrossRef] [PubMed]
- Schuster, C.F.; Mechler, L.; Nolle, N.; Krismer, B.; Zelder, M.E.; Götz, F.; Bertram, R. The MazEF Toxin-Antitoxin System Alters the β-Lactam Susceptibility of Staphylococcus aureus. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Tamber, S.; Memmi, G.; Donegan, N.P.; Cheung, A.L. Overexpression of MazFsa in Staphylococcus aureus induces bacteriostasis by selectively targeting mRNAs for cleavage. J. Bacteriol. 2009, 191, 2051–2059. [Google Scholar] [CrossRef] [PubMed]
- Yoshizumi, S.; Zhang, Y.; Yamaguchi, Y.; Chen, L.; Kreiswirth, B.N.; Inouye, M. Staphylococcus aureus YoeB homologues inhibit translation initiation. J. Bacteriol. 2009, 191, 5868–5872. [Google Scholar] [CrossRef] [PubMed]
- Larson, A.S.; Hergenrother, P.J. Light activation of Staphylococcus aureus toxin YoeBSa1 reveals guanosine-specific endoribonuclease activity. Biochemistry 2014, 53, 188–201. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.Q.; Bi, D.X.; Wei, D.Q.; Ou, H.Y. Prediction of Type II Toxin-Antitoxin Loci in Klebsiella pneumoniae Genome Sequences. Interdiscip. Sci. 2016, 8, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Tashiro, Y.; Kawata, K.; Taniuchi, A.; Kakinuma, K.; May, T.; Okabe, S. RelE-mediated dormancy is enhanced at high cell density in Escherichia coli. J. Bacteriol. 2012, 194, 1169–1176. [Google Scholar] [CrossRef] [PubMed]
- Ghafourian, S.; Good, L.; Sekawi, Z.; Hamat, R.A.; Soheili, S.; Sadeghifard, N.; Neela, V. The mazEF toxin-antitoxin system as a novel antibacterial target in Acinetobacter baumannii. Mem. Inst. Oswaldo Cruz. 2014, 109, 502–505. [Google Scholar] [CrossRef] [PubMed]
- Mosqueda, N.; Gato, E.; Roca, I.; López, M.; de Alegría, C.R.; Fernández Cuenca, F.; Martínez-Martínez, L.; Pachón, J.; Cisneros, J.M.; Rodríguez-Baño, J. Characterization of plasmids carrying the blaOXA-24/40 carbapenemase gene and the genes encoding the AbkA/AbkB proteins of a toxin/antitoxin system. J. Antimicrob. Chemother. 2014, 69, 2629–2633. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.G.; Atkinson, S.; Mathee, K.; Sam, C.K.; Chhabra, S.R.; Cámara, M.; Koh, C.L.; Williams, P. Characterization of N-acylhomoserine lactone-degrading bacteria associated with the Zingiber officinale (ginger) rhizosphere: co-existence of quorum quenching and quorum sensing in Acinetobacter and Burkholderia. BMC Microbiol. 2011, 11. [Google Scholar] [CrossRef] [PubMed]
- Bonnin, R.A.; Poirel, L.; Nordmann, P.; Eikmeyer, F.G.; Wibberg, D.; Pühler, A.; Schlüter, A. Complete sequence of broad-host-range plasmid pNOR-2000 harbouring the metallo-β-lactamase gene blaVIM-2 from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2013, 68, 1060–1065. [Google Scholar] [CrossRef] [PubMed]
- Tamman, H.; Ainelo, A.; Ainsaar, K.; Hõrak, R. A Moderate Toxin, GraT, Modulates Growth Rate and Stress Tolerance of Pseudomonas putida. J. Bacteriol. 2014, 196, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.J.; Halvorsen, E.M.; Dwyer, E.M.; DiFazio, R.M.; Hergenrother, P.J. Toxin-antitoxin (TA) systems are prevalent and transcribed in clinical isolates of Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 2011, 322, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.J.; Halvorsen, E.M.; Dwyer, E.M.; DiFazio, R.M.; Hergenrother, P.J. Toxin–antitoxin (TA) systems are prevalent and transcribed in clinical isolates of Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 2011, 322, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Wood, T.L.; Wood, T.K. The HigB/HigA toxin/antitoxin system of Pseudomonas aeruginosa influences the virulence factors pyochelin, pyocyanin, and biofilm formation. Microbiol. Open 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Ainelo, A.; Tamman, H.; Leppik, M.; Remme, J.; Hõrak, R. The toxin GraT inhibits ribosome biogenesis. Mol. Microbiol. 2016, 100, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Wood, T.K. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 2010, 391, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Dörr, T.; Vulić, M.; Lewis, K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Walker, A.N.; Daines, D.A. Toxin-antitoxin loci vapBC-1 and vapXD contribute to survival and virulence in nontypeable Haemophilus influenzae. BMC Microbial. 2012, 12. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Contreras, R.; Zhang, X.S.; Kim, Y.; Wood, T.K. Protein translation and cell death: the role of rare tRNAs in biofilm formation and in activating dormant phage killer genes. PLoS ONE 2008, 3. [Google Scholar] [CrossRef] [PubMed]
- Norton, J.P.; Mulvey, M.A. Toxin-Antitoxin Systems Are Important for Niche-Specific Colonization and Stress Resistance of Uropathogenic Escherichia coli. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Stephan, R.; Zurfluh, K.; Hächler, H.; Fanning, S. Characterization of the genetic environment of bla ESBL genes, integrons and toxin-antitoxin systems identified on large transferrable plasmids in multi-drug resistant Escherichia coli. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef]
- Schaufler, K.; Wieler, L.H.; Semmler, T.; Ewers, C.; Guenther, S. ESBL-plasmids carrying toxin-antitoxin systems can be "cured" of wild-type Escherichia coli using a heat technique. Gut Pathog. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.; Argaman, L.; Wagner, E.G.; Altuvia, S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004, 14, 2271–2276. [Google Scholar] [CrossRef] [PubMed]
- Kawano, M.; Aravind, L.; Storz, G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007, 64, 738–754. [Google Scholar] [CrossRef] [PubMed]
- Engelberg-Kulka, H.; Hazan, R.; Amitai, S. mazEF: A chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J. Cell. Sci. 2005, 118, 4327–4332. [Google Scholar] [CrossRef] [PubMed]
- Hazan, R.; Engelberg-Kulka, H. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genomics. 2004, 272, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.K.; Pedersen, K.; Hansen, F.G.; Gerdes, K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 2003, 332, 809–819. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Hara, H.; Kato, I.; Inouye, M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 2005, 280, 3143–3150. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, K.; Zavialov, A.V.; Pavlov, M.Y.; Elf, J.; Gerdes, K.; Ehrenberg, M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 2003, 112, 131–140. [Google Scholar] [CrossRef]
- Zhang, Y.; Inouye, M. The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. J. Biol. Chem. 2009, 284, 6627–6638. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kim, Y.; Hong, S.H.; Ma, Q.; Brown, B.L.; Pu, M.; Tarone, A.M.; Benedik, M.J.; Peti, W.; Page, R.; et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 2011, 7, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.Y.; Soo, V.W.; Islam, S.; McAnulty, M.J.; Benedik, M.J.; Wood, T.K. Toxin GhoT of the GhoT/GhoS toxin/antitoxin system damages the cell membrane to reduce adenosine triphosphate and to reduce growth under stress. Environ. Microbiol. 2014, 16, 1741–1754. [Google Scholar] [CrossRef] [PubMed]
- Van Acker, H.; Sass, A.; Dhondt, I.; Nelis, H.J.; Coenye, T. Involvement of toxin-antitoxin modules in Burkholderia cenocepacia biofilm persistence. Pathog. Dis. 2014, 71, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Butt, A.; Higman, V.A.; Williams, C.; mp, M.P.; Hemsley, C.M.; Harmer, N.; Titball, R.W. The HicA toxin from Burkholderia pseudomallei has a role in persister cell formation. Biochem. J. 2014, 459, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, B.; Reeves, D.S.; MacGowan, A.P. A review of the clinical presentation, laboratory features, antimicrobial therapy and outcome of 77 episodes of pneumococcal meningitis occurring in children and adults. J. Infect. 1994, 29, 171–182. [Google Scholar] [CrossRef]
- Chan, W.T.; Nieto, C.; Harikrishna, J.A.; Khoo, S.K.; Othman, R.Y.; Espinosa, M.; Yeo, C.C. Genetic regulation of the yefM-yoeB toxin-antitoxin locus of Streptococcus pneumoniae. J. Bacteriol. 2011, 193, 4612–4625. [Google Scholar] [CrossRef] [PubMed]
- Mutschler, H.; Reinstein, J.; Meinhart, A. Assembly dynamics and stability of the pneumococcal epsilon zeta antitoxin toxin (PezAT) system from Streptococcus pneumoniae. J. Biol. Chem. 2010, 285, 21797–21806. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.T.; Moreno-Cordoba, I.; Yeo, C.C.; Espinosa, M. Toxin-antitoxin genes of the Gram-positive pathogen Streptococcus pneumoniae: so few and yet so many. Microbiol. Mol. Biol. Rev. 2012, 76, 773–791. [Google Scholar] [CrossRef] [PubMed]
- Sala, A.; Bordes, P.; Genevaux, P. Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins (Basel) 2014, 6, 1002–1020. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A. Killing activity and rescue function of genome-wide toxin-antitoxin loci of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 2009, 290, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Smollett, K.L.; Fivian-Hughes, A.S.; Smith, J.E.; Chang, A.; Rao, T.; Davis, E.O. Experimental determination of translational start sites resolves uncertainties in genomic open reading frame predictions - application to Mycobacterium tuberculosis. Microbiol. 2009, 155, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Stewart, G.R.; Wernisch, L.; Stabler, R.; ngan, J.A.; Hinds, J.; Laing, K.G.; Young, D.B.; Butcher, P.D. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiol. 2002, 148, 3129–3138. [Google Scholar] [CrossRef] [PubMed]
- Tailleux, L.; Waddell, S.J.; Pelizzola, M.; Mortellaro, A.; Withers, M.; Tanne, A.; Castagnoli, P.R.; Gicquel, B.; Stoker, N.G.; Butcher, P.D.; et al. Probing host pathogen cross-talk by transcriptional profiling of both Mycobacterium tuberculosis and infected human dendritic cells and macrophages. PLoS ONE 2008, 3. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; He, Z.G. Characterization of an interplay between a Mycobacterium tuberculosis MazF homolog, Rv1495 and its sole DNA topoisomerase I. Nucleic Acids Res. 2010, 38, 8219–8230. [Google Scholar] [CrossRef] [PubMed]
- Ghafourian, S.; Raftari, M.; Sadeghifard, N.; Sekawi, Z. Toxin-antitoxin Systems: Classification, Biological Function and Application in Biotechnology. Curr. Issues Mol. Biol. 2014, 16, 9–14. [Google Scholar] [PubMed]
- Dy, R.L.; Richter, C.; Salmond, G.P.; Fineran, P.C. Remarkable Mechanisms in Microbes to Resist Phage Infections. Annu. Rev. Virol. 2014, 1, 307–331. [Google Scholar] [CrossRef] [PubMed]
- Karimi, S.; Ghafourian, S.; Taheri Kalani, M.; Azizi Jalilian, F.; Hemati, S.; Sadeghifard, N. Association between toxin-antitoxin systems and biofilm formation. Jundishapur J. Microbiol. 2015, 8. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti Infect. Ther. 2014, 12, 1221–1236. [Google Scholar] [CrossRef] [PubMed]
- Weaver, K.E.; Reddy, S.G.; Brinkman, C.L.; Patel, S.; Bayles, K.W.; Endres, J.L. Identification and characterization of a family of toxin-antitoxin systems related to the Enterococcus faecalis plasmid pAD1 par addiction module. Microbiology 2009, 155, 2930–2940. [Google Scholar] [CrossRef] [PubMed]
- Grady, R.; Hayes, F. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 2003, 47, 1419–1432. [Google Scholar] [CrossRef] [PubMed]
- Goeders, N.; Van Melderen, L. Toxin-antitoxin systems as multilevel interaction systems. Toxins (Basel) 2014, 6, 304–324. [Google Scholar] [CrossRef] [PubMed]
- Fozo, E.M.; Makarova, K.S.; Shabalina, S.A.; Yutin, N.; Koonin, E.V.; Storz, G. Abundance of type I toxin-antitoxin systems in bacteria: Searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010, 38, 3743–3759. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Inoue, K.; Yoshizumi, S.; Kobayashi, H.; Zhang, Y.; Ouyang, M.; Kato, F.; Sugai, M.; Inouye, M. Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J. Bacteriol. 2009, 191, 3248–3255. [Google Scholar] [CrossRef] [PubMed]
- Locke, J.B.; Rahawi, S.; Lamarre, J.; Mankin, A.S.; Shaw, K.J. Genetic environment and stability of cfr in methicillin-resistant Staphylococcus aureus CM05. Antimicrob. Agents Chemother. 2012, 56, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Dmowski, M.; Sitkiewicz, I.; Ceglowski, P. Characterization of a novel partition system encoded by the delta and omega genes from the streptococcal plasmid pSM19035. J. Bacteriol. 2006, 188, 4362–4372. [Google Scholar] [CrossRef] [PubMed]
- Khoo, S.K.; Loll, B.; Chan, W.T.; Shoeman, R.L.; Ngoo, L.; Yeo, C.C.; Meinhart, A. Molecular and structural characterization of the PezAT chromosomal toxin-antitoxin system of the human pathogen Streptococcus pneumoniae. J. Biol. Chem. 2007, 282, 19606–19618. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [PubMed]
- Chastre, J.; Trouillet, J.L. Problem pathogens (Pseudomonas aeruginosa and Acinetobacter). Semin. Respir. Infect. 2000, 15, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.G.; Gianoulis, T.A.; Pukatzki, S.; Mekalanos, J.J.; Ornston, L.N.; Gerstein, M.; Snyder, M. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007, 21, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Jurenaite, M.; Markuckas, A.; Suziedeliene, E. Identification and characterization of type II toxin-antitoxin systems in the opportunistic pathogen Acinetobacter baumannii. J. Bacteriol. 2013, 195, 3165–3172. [Google Scholar] [CrossRef] [PubMed]
- Lean, S.S.; Yeo, C.C.; Suhaili, Z.; Thong, K.L. Comparative Genomics of Two ST 195 Carbapenem-Resistant Acinetobacter baumannii with Different Susceptibility to Polymyxin Revealed Underlying Resistance Mechanism. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Mace, C.; Seyer, D.; Chemani, C.; Cosette, P.; Di-Martino, P.; Guery, B.; Filloux, A.; Fontaine, M.; Molle, V.; Junter, G.A.; et al. Identification of biofilm-associated cluster (bac) in Pseudomonas aeruginosa involved in biofilm formation and virulence. PLoS ONE 2008, 3. [Google Scholar] [CrossRef] [PubMed]
- Moker, N.; Dean, C.R.; Tao, J. Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J. Bacteriol. 2010, 192, 1946–1955. [Google Scholar] [CrossRef] [PubMed]
- Ryder, C.; Byrd, M.; Wozniak, D.J. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 2007, 10, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 2008, 322, 107–131. [Google Scholar] [PubMed]
- Christensen-Dalsgaard, M.; Gerdes, K. Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol. Microbiol. 2006, 62, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Budde, P.P.; Davis, B.M.; Yuan, J.; Waldor, M.K. Characterization of a higBA toxin-antitoxin locus in Vibrio cholerae. J. Bacteriol. 2007, 189, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.T.; Moreno-Cordoba, I.; Yeo, C.C.; Espinosa, M. Toxin-Antitoxin Genes of the Gram-Positive Pathogen Streptococcus pneumoniae: So Few and Yet So Many. Microbiol. Mol. Biol. Rev. 2012, 76, 773–791. [Google Scholar] [CrossRef] [PubMed]
- De la Cruz, M.A.; Zhao, W.D.; Farenc, C.; Gimenez, G.; Raoult, D.; Cambillau, C.; Gorvel, J.P.; Méresse, S. A toxin-antitoxin module of Salmonella promotes virulence in mice. PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Goulard, C.; Langrand, S.; Carniel, E.; Chauvaux, S. The Yersinia pestis chromosome encodes active addiction toxins. J. Bacteriol. 2010, 192, 3669–3677. [Google Scholar] [CrossRef] [PubMed]
- Schuessler, D.L.; Cortes, T.; Fivian-Hughes, A.S.; ougheed, K.E.; Harvey, E.; Buxton, R.S.; Davis, E.O.; Young, D.B. Induced ectopic expression of HigB toxin in Mycobacterium tuberculosis results in growth inhibition, reduced abundance of a subset of mRNAs and cleavage of tmRNA. Mol. Microbiol. 2013, 90, 195–207. [Google Scholar] [PubMed]
- Pandey, D.P.; Gerdes, K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005, 33, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Christensen-Dalsgaard, M.; Jorgensen, M.G.; Gerdes, K. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 2010, 75, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.M.; Woychik, N.A. Bacterial Toxin HigB Associates with Ribosomes and Mediates Translation-dependent mRNA Cleavage at A-rich Sites. J. Biol. Chem. 2009, 284, 18605–18613. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.R.; Burns, J.L.; Lory, S.; Lewis, K. Emergence of Pseudomonas aeruginosa Strains Producing High Levels of Persister Cells in Patients with Cystic Fibrosis. J. Bacteriol. 2010, 192, 6191–6199. [Google Scholar] [CrossRef] [PubMed]
- Kwan, B.W.; Valenta, J.A.; Benedik, M.J.; Wood, T.K. Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother. 2013, 57, 1468–1473. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister cells, dormancy and infectious disease. Nat. Rev. Microb. 2007, 5, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Wood, T.K. Combatting bacterial persister cells. Biotechnol. Bioeng. 2016, 113, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wood, T.K. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 2011, 77, 5577–5583. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.; Zhang, Z.; Khodursky, A.; Kaldalu, N.; Kurg, K.; Lewis, K. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006, 6. [Google Scholar] [CrossRef] [PubMed]
- Keren, I.; Kaldalu, N.; Spoering, A.; Wang, Y.; Lewis, K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 2004, 230, 13–18. [Google Scholar] [CrossRef]
- (CDC) CfDC. Isolation of E. coli O157:H7 from sporadic cases of hemorrhagic colitis - United States. MMWR Morb. Mortal. Wkly. Rep. 1982, 31, 580–585. [Google Scholar]
- Hazan, R.; Sat, B.; Engelberg-Kulka, H. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 2004, 186, 3663–3669. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Bedzyk, L.A.; Thomas, S.M.; Ye, R.W.; Wood, T.K. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 2004, 64, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.L.; Grigoriu, S.; Kim, Y.; rruda, J.M.; Davenport, A.; Wood, T.K.; Peti, W.; Page, R. Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 2009, 5. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.L.; Wood, T.K.; Peti, W.; Page, R. Structure of the Escherichia coli antitoxin MqsA (YgiT/b3021) bound to its gene promoter reveals extensive domain rearrangements and the specificity of transcriptional regulation. J. Biol. Chem. 2011, 286, 2285–2296. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lord, D.M.; Hong, S.H.; Peti, W.; Benedik, M.J.; Page, R.; Wood, T.K. Type II toxin/antitoxin MqsR/MqsA controls type V toxin/antitoxin GhoT/GhoS. Environ. Microbiol. 2013, 15, 1734–1744. [Google Scholar] [CrossRef] [PubMed]
- Kwan, B.W.; Lord, D.M.; Peti, W.; Page, R.; Benedik, M.J.; Wood, T.K. The MqsR/MqsA toxin/antitoxin system protects Escherichia coli during bile acid stress. Environ. Microbiol. 2015, 17, 3168–3181. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Park, J.H.; Inouye, M. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J. Biol. Chem. 2009, 284, 28746–28753. [Google Scholar] [CrossRef] [PubMed]
- Lipuma, J.J. Update on the Burkholderia cepacia complex. Curr. Opin. Pulm. Med. 2005, 11, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Mahenthiralingam, E.; Urban, T.A.; Goldberg, J.B. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 2005, 3, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Currie, B.J.; Fisher, D.A.; Anstey, N.M.; Jacups, S.P. Melioidosis: acute and chronic disease, relapse and re-activation. Trans. R. Soc. Trop. Med. Hyg. 2000, 94, 301–304. [Google Scholar] [CrossRef]
- Agnoli, K.; Frauenknecht, C.; Freitag, R.; hwager, S.; Jenul, C.; Vergunst, A.; Carlier, A.; Eberl, L. The third replicon of members of the Burkholderia cepacia Complex, plasmid pC3, plays a role in stress tolerance. Appl. Environ. Microbiol. 2014, 80, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Nieto, C.; Cherny, I.; Khoo, S.K.; e Lacoba, M.G.; Chan, W.T.; Yeo, C.C.; Gazit, E.; Espinosa, M. The yefM-yoeB toxin-antitoxin systems of Escherichia coli and Streptococcus pneumoniae: Functional and structural correlation. J. Bacteriol. 2007, 189, 1266–1278. [Google Scholar] [CrossRef] [PubMed]
- Behnke, D.; Malke, H.; Hartmann, M.; Walter, F. Post-transformational rearrangement of an in vitro reconstructed group-A streptococcal erythromycin resistance plasmid. Plasmid 1979, 2, 605–616. [Google Scholar] [CrossRef]
- Ceglowski, P.; Boitsov, A.; Karamyan, N.; Chai, S.; Alonso, J.C. Characterization of the effectors required for stable inheritance of Streptococcus pyogenes pSM19035-derived plasmids in Bacillus subtilis. Mol. Gen. Genet. 1993, 241, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Nieto, C.; Sadowy, E.; de la Campa, A.G.; Hryniewicz, W.; Espinosa, M. The relBE2Spn toxin-antitoxin system of Streptococcus pneumoniae: Role in antibiotic tolerance and functional conservation in clinical isolates. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, W.T.; Yeo, C.C.; Sadowy, E.; Espinosa, M. Functional validation of putative toxin-antitoxin genes from the Gram-positive pathogen Streptococcus pneumoniae: phd-doc is the fourth bona-fide operon. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Tuberculosis. Available online: http://www.who.int/mediacentre/factsheets/fs104/en/ (accessed on 10 June 2016).
- Han, J.S.; Lee, J.J.; Anandan, T.; Sripathi, S.; Jahng, W.J.; Lee, S.H.; Suh, J.W.; Kang, C.M. Characterization of a chromosomal toxin-antitoxin, Rv1102c-Rv1103c system in Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2010, 400, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Barry, C.E.; Boshoff, H.I. The three RelE homologs of Mycobacterium tuberculosis have individual, drug-specific effects on bacterial antibiotic tolerance. J. Bacteriol. 2010, 192, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Korch, S.B.; Contreras, H.; Clark-Curtiss, J.E. Three Mycobacterium tuberculosis Rel toxin-antitoxin modules inhibit mycobacterial growth and are expressed in infected human macrophages. J. Bacteriol. 2009, 191, 1618–1630. [Google Scholar] [CrossRef] [PubMed]
- Keren, I.; Minami, S.; Rubin, E.; Lewis, K. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. MBio 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Stinear, T.P.; Seemann, T.; Harrison, P.F.; Jenkin, G.A.; Davies, J.K.; Johnson, P.D.; Abdellah, Z.; Arrowsmith, C.; Chillingworth, T.; Churcher, C.; et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008, 18, 729–741. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Sharp, J.D.; Kobayashi, H.; Woychik, N.A.; Inouye, M. Noncognate Mycobacterium tuberculosis toxin-antitoxins can physically and functionally interact. J. Biol. Chem. 2010, 285, 39732–39738. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.G.; Lee, S.J.; Chae, S.; Lee, K.Y.; Kim, J.H.; Lee, B.J. Structural and functional studies of the Mycobacterium tuberculosis VapBC30 toxin-antitoxin system: implications for the design of novel antimicrobial peptides. Nucleic Acids Res. 2015, 43, 7624–7637. [Google Scholar] [CrossRef] [PubMed]
- Albrethsen, J.; Agner, J.; Piersma, S.R.; Højrup, P.; Pham, T.V.; Weldingh, K.; Jimenez, C.R.; Andersen, P.; Rosenkrands, I. Proteomic profiling of Mycobacterium tuberculosis identifies nutrient-starvation-responsive toxin-antitoxin systems. Mol. Cell. Proteom. 2013, 12, 1180–1191. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Gonnet, M.; Hania, W.B.; Forterre, P.; Erauso, G. Insights into dynamics of mobile genetic elements in hyperthermophilic environments from five new Thermococcus plasmids. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wibberg, D.; Szczepanowski, R.; Eikmeyer, F.; Puhler, A.; Schluter, A. The IncF plasmid pRSB225 isolated from a municipal wastewater treatment plant's on-site preflooder combining antibiotic resistance and putative virulence functions is highly related to virulence plasmids identified in pathogenic E. coli isolates. Plasmid 2013, 69, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Saluja, S.K.; Weiser, J.N. The genetic basis of colony opacity in Streptococcus pneumoniae: Evidence for the effect of box elements on the frequency of phenotypic variation. Mol. Microbiol. 1995, 16, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Arora, G.; Singh, M.; Kidwai, S.; Narayan, O.P.; Singh, R. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Helaine, S.; Cheverton, A.M.; Watson, K.G.; Faure, L.M.; Matthews, S.A.; Holden, D.W. Internalization of Salmonella by Macrophages Induces Formation of Nonreplicating Persisters. Science 2014, 343, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.A.; Kriegeskorte, A.; Kahl, B.C.; Becker, K.; Löffler, B.; Peters, G. Staphylococcus aureus Small Colony Variants (SCVs): A road map for the metabolic pathways involved in persistent infections. Front. Cell. Infect. Microbiol. 2014, 4, 99. [Google Scholar] [PubMed]
- Conlon, B.P. Staphylococcus aureus chronic and relapsing infections: Evidence of a role for persister cells: An investigation of persister cells, their formation and their role in S. aureus disease. Bioessays 2014, 36, 991–996. [Google Scholar] [CrossRef] [PubMed]
- Gelens, L.; Hill, L.; Vandervelde, A.; Danckaert, J.; Loris, R. A general model for toxin-antitoxin module dynamics can explain persister cell formation in E. coli. PLoS Comput. Biol. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Pecota, D.C.; Wood, T.K. Exclusion of T4 Phage by the hok/sok Killer Locus from Plasmid R1. J. Bacteriol. 1996, 178, 2044–2050. [Google Scholar] [PubMed]
- Fineran, P.C.; Blower, T.R.; Foulds, I.J.; Humphreys, D.P.; Lilley, K.S.; Salmond, G.P. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. USA 2009, 106, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Blower, T.R.; Short, F.L.; Rao, F.; guchi, K.; Pei, X.Y.; Fineran, P.C.; Luisi, B.F.; Salmond, G.P. Identification and classification of bacterial Type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res. 2012, 40, 6158–6173. [Google Scholar] [CrossRef] [PubMed]
- Samson, J.E.; Spinelli, S.; Cambillau, C.; Moineau, S. Structure and activity of AbiQ, a lactococcal endoribonuclease belonging to the type III toxin-antitoxin system. Mol. Microbiol. 2013, 87, 756–768. [Google Scholar] [CrossRef] [PubMed]
- Anukam, K.C.; Macklaim, J.M.; Gloor, G.B.; Reid, G.; Boekhorst, J.; Renckens, B.; van Hijum, S.A.; Siezen, R.J. Genome sequence of Lactobacillus pentosus KCA1: Vaginal isolate from a healthy premenopausal woman. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dy, R.L.; Przybilski, R.; Semeijn, K.; Salmond, G.P.; Fineran, P.C. A widespread bacteriophage abortive infection system functions through a Type IV toxin-antitoxin mechanism. Nucleic Acids Res. 2014, 42, 4590–4605. [Google Scholar] [CrossRef] [PubMed]
- Fiedoruk, K.; Daniluk, T.; Swiecicka, I.; Sciepuk, M.; Leszczynska, K. Type II toxin-antitoxin systems are unevenly distributed among Escherichia coli phylogroups. Microbiology 2015, 161, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Rasko, D.A.; Rosovitz, M.J.; Myers, G.S.; Mongodin, E.F.; Fricke, W.F.; Gajer, P.; Crabtree, J.; Sebaihia, M.; Thomson, N.R.; Chaudhuri, R.; et al. The pangenome structure of Escherichia coli: Comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 2008, 190, 6881–6893. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, N.; Kwan, B.W.; Wood, T.K. Persistence Increases in the Absence of the Alarmone Guanosine Tetraphosphate by Reducing Cell Growth. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]


| Bacterium | TA System | Type | Localization | Function | Other Pathogens | References |
|---|---|---|---|---|---|---|
| ESKAPE Group | ||||||
| Enterococcus spp. | Par locus | I | Plasmid pAD | Regulation, persistence and plasmid maintenance | Lactobacillus casei S. aureus | [20,21] |
| Axe/Txe | II | Plasmid pRUM | Plasmid maintenance and vancomycin resistance (VanA enzyme) | S. aureus E. coli | [22] | |
| Omega/Epsilon/Zeta | II | Plasmid pVEF3 | Plasmid maintenance vancomycin resistance (VanA enzyme) | Bacillus subtilis E. coli S. aureus | [23,24] | |
| HigBA | II | Chromosome | Expression of virulence factors | Enterococcus spp. Proteus vulgaris Vibrio cholerae E. coli S. pneumoniae A. baumannii P. aeruginosa Salmonella typhimurium Yersinia pestis M. tuberculosis | [25] | |
| MazEF | II | Chromosome | Expression of virulence factors | S. aureus E. coli S. typhimurium P. aeruginosa M. Tuberculosis | [25] | |
| S.aureus | PemIK | II | Plasmid cCHP91 | Plasmid maintenance and global regulation of virulence | - | [26] |
| SprG1/Spr1 | II | Phage | Lytic activity (human erythrocytes) | - | [27,28] | |
| SprA1 (PepA1/AS) | I | Pathogenicityisland/Chromosome | Persistence and pathogenicity | - | [29] | |
| MazEF | II | Chromosome | Regulation of β-lactamase sensitivity | Enterococcus spp. E. coli P. aeruginosa S. pneumoniae M. tuberculosis | [30,31,32] | |
| YefM/YoeB | II | Chromosome | Cell arrest | - | [33,34] | |
| Omega/Epsilon/Zeta | Chromosome | - | Bacillus subtilis E. coli | [29] | ||
| K. pneumoniae | RelBE_1 | II | Plasmid | Persistence of cells against antibiotics and plasmid maintenance | A. baumannii P. aeruginosa E. coli B. pseudomallei S. pneumoniae | [35,36] |
| RelBE_2 | II | Chromosome | M. tuberculosis | |||
| A. baumannii | RelBE, HigBA, SplTA and CheTA | II | Plasmid p3ABAYE | Plasmid maintenance | Enterococcus spp. Proteus vulgaris Vibrio cholerae E. coli S. pneumoniae P. aeruginosa Salmonella typhimurium Yersinia pestis M. tuberculosis | [37] |
| AbKAB (SplTA) | II | Plasmid | Plasmid maintenance and carbapenem resistance (OXA 24 ß-lactamase) | E. coli | [38] | |
| GraTA | II | Plasmid | - | P. aeruginosa | [39] | |
| MazEF | II | Chromosome | - | Enterococcus spp. S. aureus P. aeruginosa E. coli S. typhimurium M. tuberculosis | [37,39] | |
| P.aeruginosa | ParAB, TOX1/TOX2, T/AT1-2 | II | Plasmid pNOR-2000 | Plasmid maintenance and carbapenem resistance(VIM metallo-ß-lactamase) | Enterococcus spp. Lactobacillus casei S. aureus | [40] |
| RelBE | II | Chromosome | - | K. pneumoniae A. baumannii E. coli B. pseudomallei S. pneumoniae M. tuberculosis | [41,42,43] | |
| HigBA | II | Chromosome/ Plasmid Rts1 | Reduction of pyochelin, swarming and biofilm formation | Enterococcus spp. P. vulgaris V. cholerae S. pneumoniae A. baumannii E. coli S. typhimurium Yersinia pestis M. tuberculosis E. coli | [42,44] | |
| GraTA | II | Chromosome | Persistence | A. baumannii | [45] | |
| MazEF | II | Chromosome | Persistence | Enterococcus spp. S. aureus E. coli S. typhimurium M. tuberculosis | [46,47] | |
| Vap-type systems | II | - | Regulation of virulence | Haemophilus influenzae | [48] | |
| YefM/YoeB | II | Chromosome | Regulation of virulence | E. coli S. aureus S. pneumoniae M. tuberculosis | [49] | |
| Hha/TomB | II | Chromosome | Regulation of virulence | E. coli | [49] | |
| PasTI | II | Chromosome | Regulation of virulence | E. coli | [50] | |
| Enterobacter spp. | - | - | - | - | - | |
| Other pathogens | ||||||
| E.coli | PemIK, VagCD, CcdAB, Hok/Sok, ParAB and PsiAB | II | Plasmid pEC302104 | Plasmid maintenance and ß-lactam resistance (ESBL ß-lactamase) | Enterococcus spp. S. aureus | [51,52] |
| TisB/IstR | I | Chromosome | Regulation of SOS response | - | [4,53] | |
| SymER | I | Chromosome | Regulation of SOS response | - | [54] | |
| MazEF | II | Chromosome | Persistence, biofilm formation | Enterococcus spp. S. aureus S. typhimurium P. aeruginosa M. tuberculosis | [55,56,57,58] | |
| RelBE | II | Chromosome | Persistence, biofilm formation | K. pneumoniae A. baumannii P. aeruginosa B. pseudomallei S. pneumoniae M. tuberculosis | [59] | |
| YefM/YoeB | II | Chromosome | Persistence, biofilm formation | S. aureus, P. aeruginosa S. pneumoniae | [60] | |
| MqsRA | II | Chromosome | Influence on biofilm formation and global stress response. Control of GhoTS System. Increased tolerance to bile acid. | - | [49,50,51,52,53,54,55,56,57,58,59,60,61] | |
| GhoTS | V | Chromosome | Persistence, biofilm formation | - | [49,62] | |
| Hha/TomB | II | Chromosome | Persistence, decreases biofilm formation by inhibiting fimbriae production. | P. aeruginosa | [49,61] | |
| PasTI | II | - | Persistence | P. aeruginosa | [50] | |
| Burkholderia spp. | TAS1/TAS2 | II | Plasmid pC3 | Plasmid maintenance and tolerance to antibiotics | - | [63,64] |
| RelBE | II | Chromosome | Persistence | K. pneumoniae A. baumannii P. aeruginosa E. coli S. pneumoniae M. tuberculosis | [63] | |
| HicAB | II | Chromosome | Persistence | E. coli | [64,65] | |
| Streptococcus spp. | YefM/YoeB | II | - | Implicated in pathogenicity, phase variation, genetic competence, biofilm formation and bistability | E. coli S. aureus P. aeruginosa M. tuberculosis | [66,67] |
| PezAT | II | - | Persistence and biofilm formation | - | ||
| RelBE | II | Chromosome | Associated with survival and human colonization | K. pneumoniae A. baumannii P. aeruginosa E. coli B. pseudomallei M. tuberculosis | [66] | |
| Phd-Doc | - | - | - | [68] | ||
| M.tuberculosis | YefM/YoeB | II | Chromosome | Persistence | E. coli S. aureus P. aeruginosa S. pneumoniae | [69,70,71] |
| RelBE | II | Chromosome | Persistence | K. pneumoniae A. baumannii P. aeruginosa E. coli B. pseudomallei S. pneumoniae | [70,71] | |
| ParDE | II | Chromosome | Persistence | - | [72] | |
| HigBA | II | Chromosome | Persistence | Enterococcus spp. P. vulgaris V. cholerae A. baumannii E. coli S. pneumoniae Salmonella typhimurium Yersinia pestis | [73,74] | |
| TAC | Chromosome | - | - | [69] | ||
| MazEF | II | Chromosome | Persistence and cell arrest | Enterococcus spp. S. aureus E. coli S. typhimurium P. aeruginosa | [75] | |
| VapBC | II | Chromosome | Persistence | P. aeruginosa H. influenzae | [17,76,77] | |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-García, L.; Blasco, L.; Lopez, M.; Bou, G.; García-Contreras, R.; Wood, T.; Tomas, M. Toxin-Antitoxin Systems in Clinical Pathogens. Toxins 2016, 8, 227. https://doi.org/10.3390/toxins8070227
Fernández-García L, Blasco L, Lopez M, Bou G, García-Contreras R, Wood T, Tomas M. Toxin-Antitoxin Systems in Clinical Pathogens. Toxins. 2016; 8(7):227. https://doi.org/10.3390/toxins8070227
Chicago/Turabian StyleFernández-García, Laura, Lucia Blasco, Maria Lopez, German Bou, Rodolfo García-Contreras, Thomas Wood, and María Tomas. 2016. "Toxin-Antitoxin Systems in Clinical Pathogens" Toxins 8, no. 7: 227. https://doi.org/10.3390/toxins8070227
APA StyleFernández-García, L., Blasco, L., Lopez, M., Bou, G., García-Contreras, R., Wood, T., & Tomas, M. (2016). Toxin-Antitoxin Systems in Clinical Pathogens. Toxins, 8(7), 227. https://doi.org/10.3390/toxins8070227