Palladium Nanoparticles-Based Fluorescence Resonance Energy Transfer Aptasensor for Highly Sensitive Detection of Aflatoxin M1 in Milk
Abstract
:1. Introduction
2. Results and Discussion
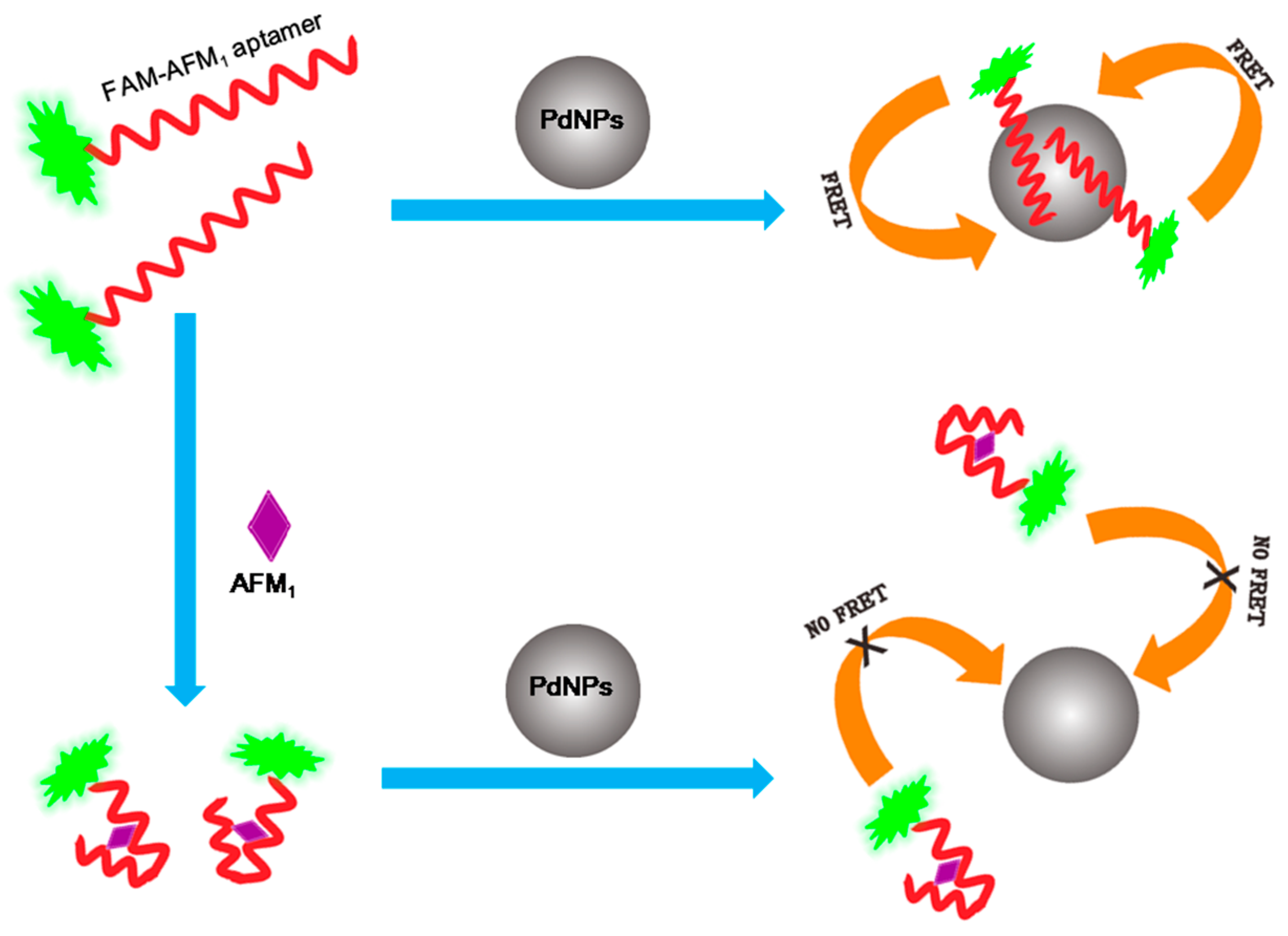
2.1. Construction of the FRET Aptasensor for AFM1
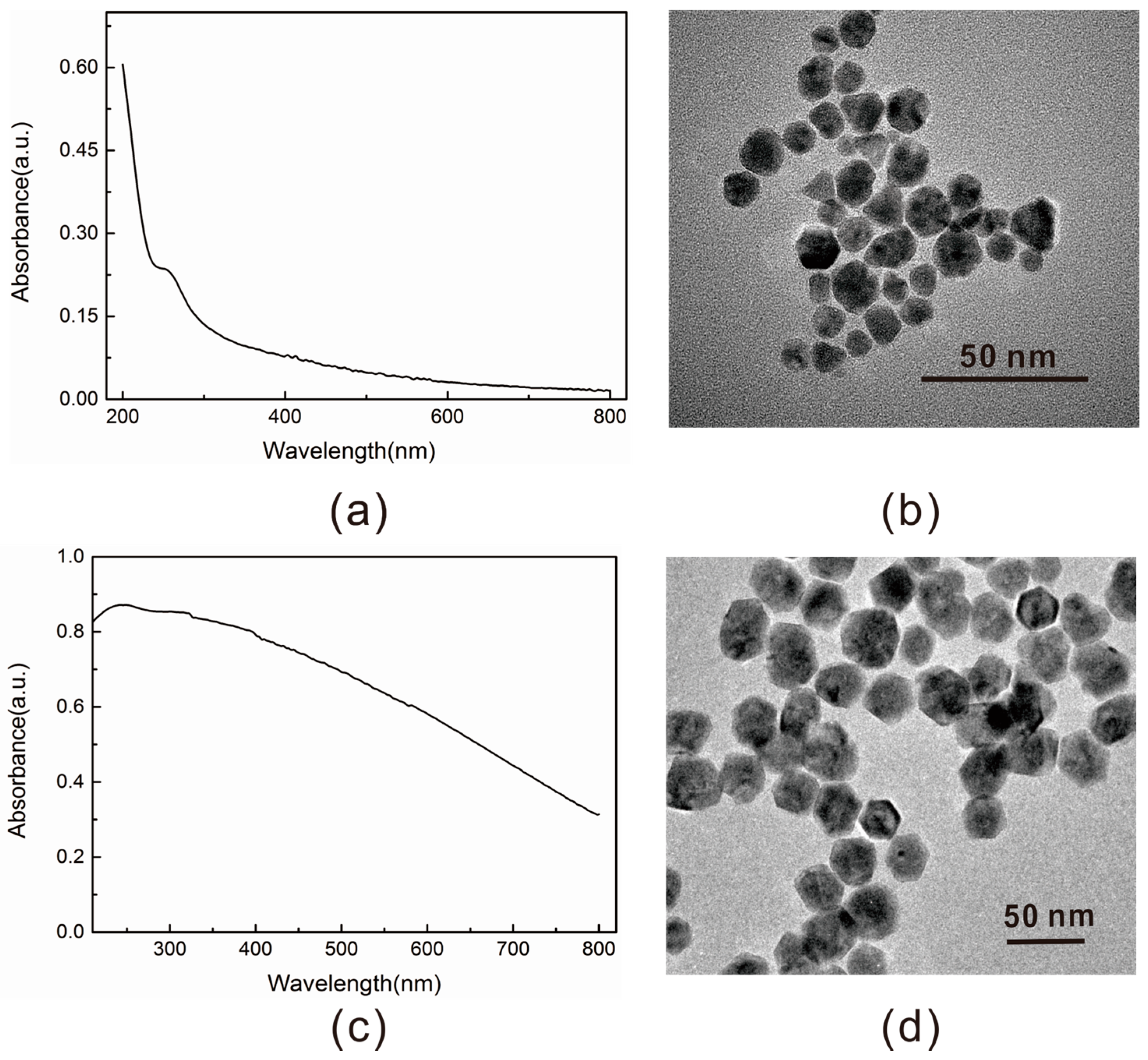
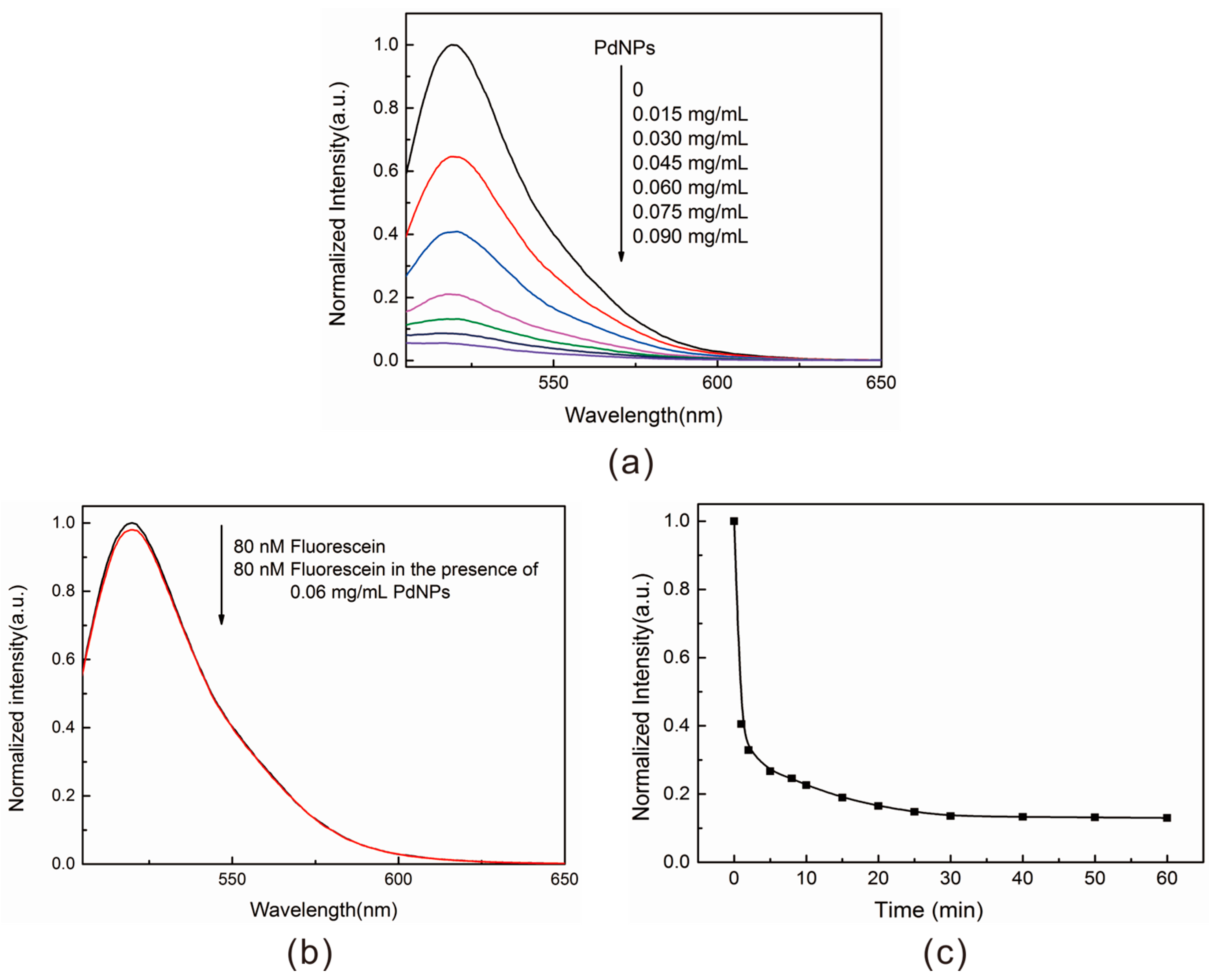
2.2. Properties Characterization of the Energy Acceptor
2.3. Construction of the AFM1 Aptasensor
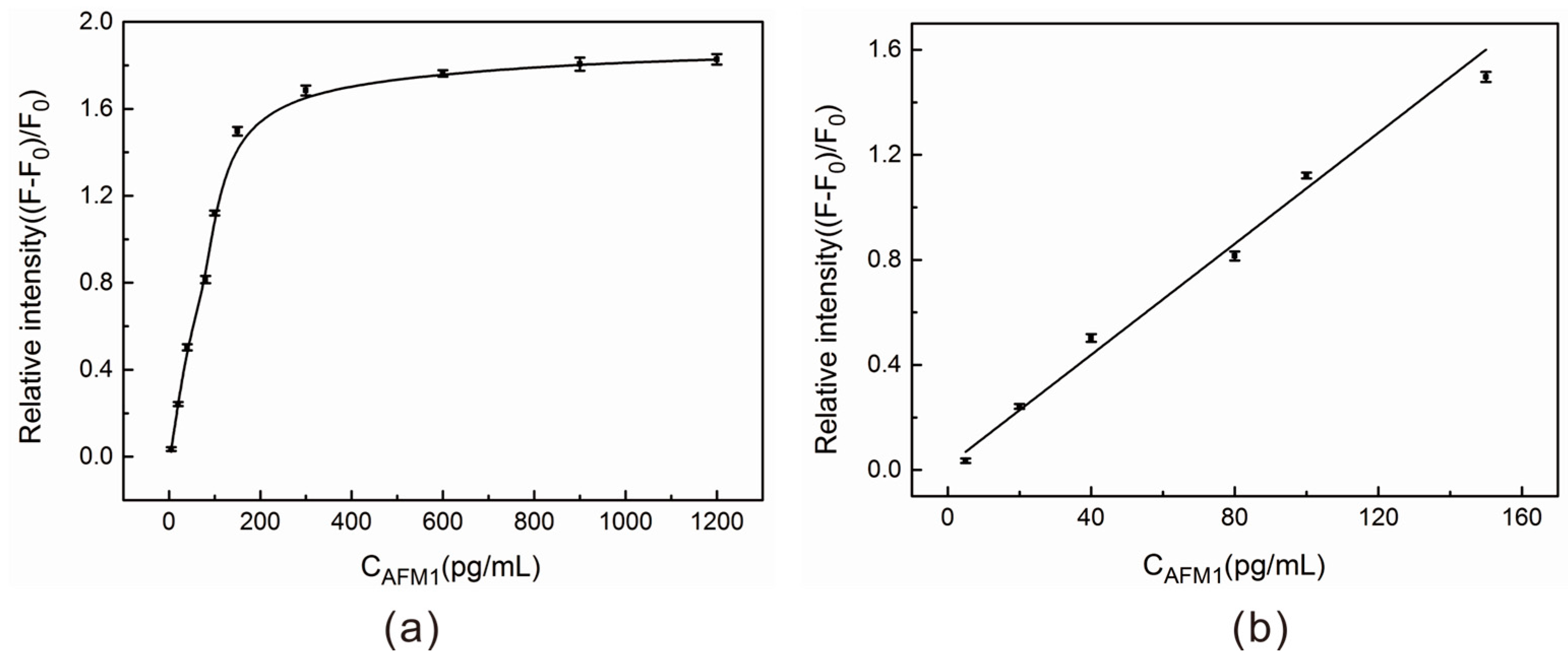
2.4. AFM1 Detection in Aqueous Buffer Solution
2.5. AFM1 Detection in Milk Samples
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Instrumentation
4.3. Synthesis of Sodium Citrate Capped Palladium Seeds
4.4. The Synthesis of 33 nm PdNPs
4.5. Quenching Measurements
4.6. AFM1 Detection in Aqueous Buffer Solution
4.7. AFM1 Detection in Milk Samples
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Torres, A.M.; Barros, G.G.; Palacios, S.A.; Chulze, S.N.; Battilani, P. Review on pre- and post-harvest management of peanuts to minimize aflatoxin contamination. Food Res. Int. 2014, 62, 11–19. [Google Scholar] [CrossRef]
- Bakirci, I. A study on the occurrence of aflatoxin M1 in milk and milk products produced in Van province of Turkey. Food Control 2001, 12, 47–51. [Google Scholar] [CrossRef]
- Richard, J.L. Some major mycotoxins and their mycotoxicoses-an overview. Int. J. Food Microbiol. 2007, 119, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Kanungo, L.; Pal, S.; Bhand, S. Miniaturised hybrid immunoassay for high sensitivity analysis of aflatoxin M1 in milk. Biosens. Bioelectron. 2011, 26, 2601–2606. [Google Scholar] [CrossRef] [PubMed]
- International Agency for Research on Cancer. Monograph on the Evaluation of Carcinogenic Risks in Humans: Some Traditional Herbal Medicines, Some Mycotoxins; IARC: Lyon, France, 2002; pp. 171–274. [Google Scholar]
- Iha, M.H.; Barbosa, C.B.; Okada, I.A.; Trucksess, M.W. Aflatoxin M1 in milk and distribution and stability of aflatoxin M1 during production and storage of yoghurt and cheese. Food Control 2013, 29, 1–6. [Google Scholar] [CrossRef]
- Badea, M.; Micheli, L.; Messia, M.C.; Candigliota, T.; Marconi, E.; Mottram, T.; Velasco-Garcia, M.; Moscone, D.; Palleschi, G. Aflatoxin M1 determination in raw milk using a flow-injection immunoassay system. Anal. Chim. Acta 2004, 520, 141–148. [Google Scholar] [CrossRef] [Green Version]
- European Commission, Commission Regulation (EC) No 1881/2006 of 19 December 2006 setting maximum levels for certain contaminants in foodstuffs. Off. J. Eur. Union 2006, 364, 5–24.
- US Food and Drug Administration. 400 Whole Milk, Low Fat Milk, Skim Milk-Aflatoxin M1 (cpg 7106.210), FDA Compliance Policy Guides; FDA: Washington, DC, USA, 2005; p. 219.
- MoH (Ministry of Health) P.R. China. Maximum Residue Level of Mycotoxin in Food-National Regulations for Food Safety; National Standard No. 2761-2011; Ministry of Health: Beijing, China, 2011.
- Lin, L.M.; Zhang, J.; Wang, P.; Wang, Y.S.; Chen, J.P. Thin-layer chromatography of mycotoxins and comparison with other chromatographic methods. J. Chromatogr. A 1998, 815, 3–20. [Google Scholar] [CrossRef]
- Filazi, A.; Ince, S.; Temamogullari, F. Survey of the occurrence of aflatoxin M1 in cheeses produced by dairy ewe’s milk in Urfa city, Turkey. Ankara Üniv. Vet. Fak. Derg. 2010, 57, 197–199. [Google Scholar] [CrossRef]
- Anfossi, L.; Calderara, M.; Baggiani, C.; Giovannoli, C.; Arletti, E.; Giraudi, G. Development and application of solvent-free extraction for the detection of aflatoxin M1 in dairy products by enzyme immunoassay. J. Agric. Food Chem. 2008, 56, 1852–1857. [Google Scholar] [CrossRef] [PubMed]
- Chadseesuwan, U.; Sangdokmai, A.; Pimpitak, U.; Puthong, S.; Palaga, T.; Komolpis, K. Production of a monoclonal antibody against aflatoxin M1 and its application for detection of aflatoxin M1 in fortified milk. J. Food Drug Anal. 2016, 24, 780–787. [Google Scholar] [CrossRef] [PubMed]
- Shuib, N.S.; Makahleh, A.; Salhimi, S.M.; Saad, B. Determination of aflatoxin M1 in milk and dairy products using high performance liquid chromatography-fluorescence with post column photochemical derivatization. J. Chromatogr. A 2017, 1510, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, M.; Taherimaslak, Z. Determination of aflatoxin M-1 in liquid milk using high performance liquid chromatography with fluorescence detection after magnetic solid phase extraction. RSC Adv. 2014, 4, 33497–33506. [Google Scholar] [CrossRef]
- Huang, S.M.; Hu, D.; Wang, Y.; Zhu, F.; Jiang, R.F.; Ouyang, G.F. Automated hollow-fiber liquid-phase microextraction coupled with liquid chromatography/tandem mass spectrometry for the analysis of alfatoxin M-1 in milk. J. Chromatogr. A 2015, 1416, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Campone, L.; Piccinelli, A.L.; Celano, R.; Russo, M.; Rastrelli, L. Rapid analysis of aflatoxin M-1 in milk using dispersive liquid-liquid microextraction coupled with ultrahigh pressure liquid chromatography tandem mass spectrometry. Anal. Bioanal. Chem. 2013, 405, 8645–8652. [Google Scholar] [CrossRef] [PubMed]
- Parker, C.O.; Lanyon, Y.H.; Manning, M.; Arrigan, D.W.M.; Tothill, I.E. Electrochemical immunochip sensor for aflatoxin M1 detection. Anal. Chem. 2009, 81, 5291–5298. [Google Scholar] [CrossRef] [PubMed]
- Gan, N.; Zhou, J.; Xiong, P.; Hu, F.T.; Cao, Y.T.; Li, T.H.; Jiang, Q.L. An ultrasensitive electrochemiluminescent immunoassay for aflatoxin M1 in milk, based on extraction by magnetic graphene and detection by antibody-labeled CdTe quantum dots-carbon nanotubes nanocomposite. Toxins 2013, 5, 865–883. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.L.; Yang, S.M. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens. Bioelectron. 2015, 71, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Dinçkaya, E.; Kınık, Ö.; Sezgintürk, M.K.; Altuğ, Ҫ.; Akkoca, A. Development of an impedimetric aflatoxin M1 biosensor based on a DNA probe and gold nanoparticles. Biosens. Bioelectron. 2011, 26, 3806–3811. [Google Scholar] [CrossRef]
- Nguyen, B.H.; Tran, L.D.; Do, Q.P.; Nguyen, H.L.; Tran, N.H.; Nguyen, P.X. Label-free detection of aflatoxin M1 with electrochemical Fe3O4/polyaniline-based aptasensor. Mater. Sci. Eng. C 2013, 33, 2229–2234. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Zhao, M.; Zhuo, Y.; Chai, Y.Q.; Yuan, R. Highly efficient intramolecular electrochemiluminescence energy transfer for ultrasensitive bioanalysis of alfatoxin M1. Chem. Eur. J. 2017, 23, 1853–1859. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Catanante, G.; Hayat, A.; Istamboulie, G.; Rejeb, I.B.; Bhand, S.; Marty, J.L. Development of structure switching aptamer assay for detection of aflatoxin M1 in milk sample. Talanta 2016, 158, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Wegner, K.D.; Lindén, S.; Jin, Z.; Jennings, T.L.; Khoulati, R.; van Bergen en Henegouwen, P.M.P.; Hildebrandt, N. Nanobodies and nanocrystals: Highly sensitive quantum dot-based homogeneous FRET immunoassay for serum-based EGFR detection. Small 2014, 10, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Lao, Y.H.; Chi, C.W.; Friedrich, S.M.; Peck, K.; Wang, T.H.; Leong, K.W.; Chen, L.C. Signal-on protein detection via dye translocation between aptamer and quantum dot. ACS Appl. Mater. Interfaces 2016, 8, 12048–12055. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Jiang, K.; Zhu, J.L.; Zhang, L.; Lin, H.W. A FRET-based carbon dot-MnO2 nanosheet architecture for glutathione sensing in human whole blood samples. Chem. Commun. 2015, 51, 12748–12751. [Google Scholar] [CrossRef] [PubMed]
- Arola, H.O.; Tullila, A.; Kiljunen, H.; Campbell, K.; Siitari, H.; Nevanen, T.K. Specific noncompetitive immunoassay for HT-2 mycotoxin detection. Anal. Chem. 2016, 88, 2446–2452. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Z.; Jin, Y.; Wang, L.; Deng, J.; Zhang, C.X. Gold nanorods-based FRET assay for ultrasensitive detection of Hg2+. Chem. Commun. 2011, 47, 12500–12502. [Google Scholar] [CrossRef] [PubMed]
- Maity, D.; Karthigeyan, D.; Kundu, T.K.; Govindaraju, T. FRET-based rational strategy for ratiometric detetction of Cu2+ and live cell imaging. Sensor Actuators B Chem. 2013, 176, 831–837. [Google Scholar] [CrossRef]
- Li, H.; Sun, D.E.; Liu, Z.H. Ultrasensitive biosensing platform based on the luminescence quenching ability of plasmonic palladium nanoparticles. Chem. Eur. J. 2015, 21, 4944–4948. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Shi, L.; Sun, D.E.; Li, P.W.; Liu, Z.H. Fluorescence resonance energy transfer biosensor between upconverting nanoparticles and palladium nanoparticles for ultrasensitive CEA detection. Biosens. Bioelectron. 2016, 86, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.Q.; Zhang, Z.W.; Li, P.W.; Zhang, Q.; Jiang, J.; Wang, D.; Lei, J.W. Sample-pretreatment-free based high sensitive determination of aflatoxin M1 in raw milk using a time-resolved fluorescent competitive immunochromatographic assay. RSC Adv. 2015, 5, 558–564. [Google Scholar] [CrossRef]
- Meyer, D.A.; Albrecht, R.M. Sodium ascorbate method for the synthesis of colloidal palladium particles of different sizes. Microsc. Microanal. 2003, 9, 1190–1191. [Google Scholar] [CrossRef]
- Lu, L.H.; Wang, H.S.; Xi, S.Q.; Zhang, H.J. Improved size control of large palladium nanoparticles by a seeding growth method. J. Mater. Chem. 2002, 12, 156–158. [Google Scholar] [CrossRef]
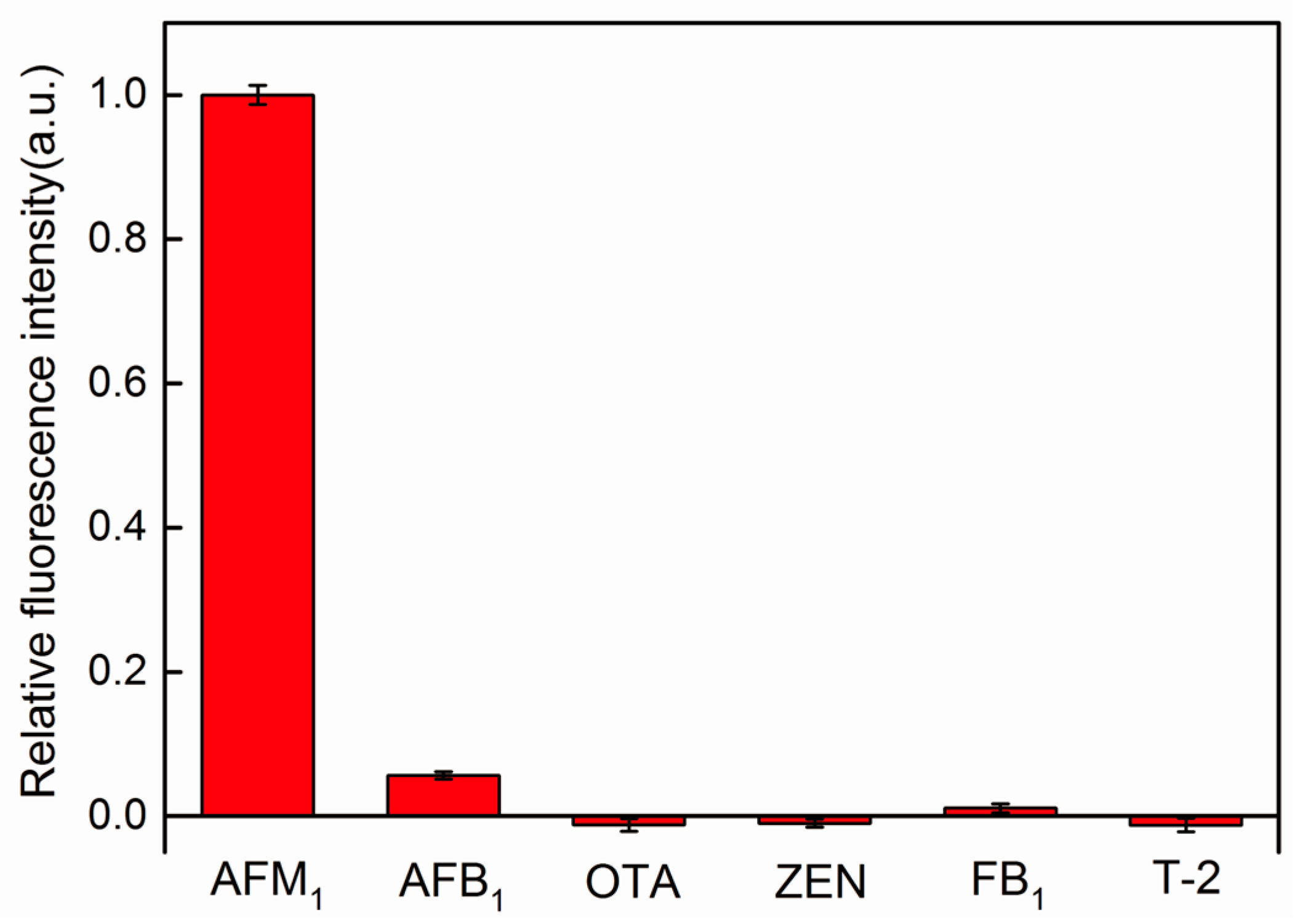
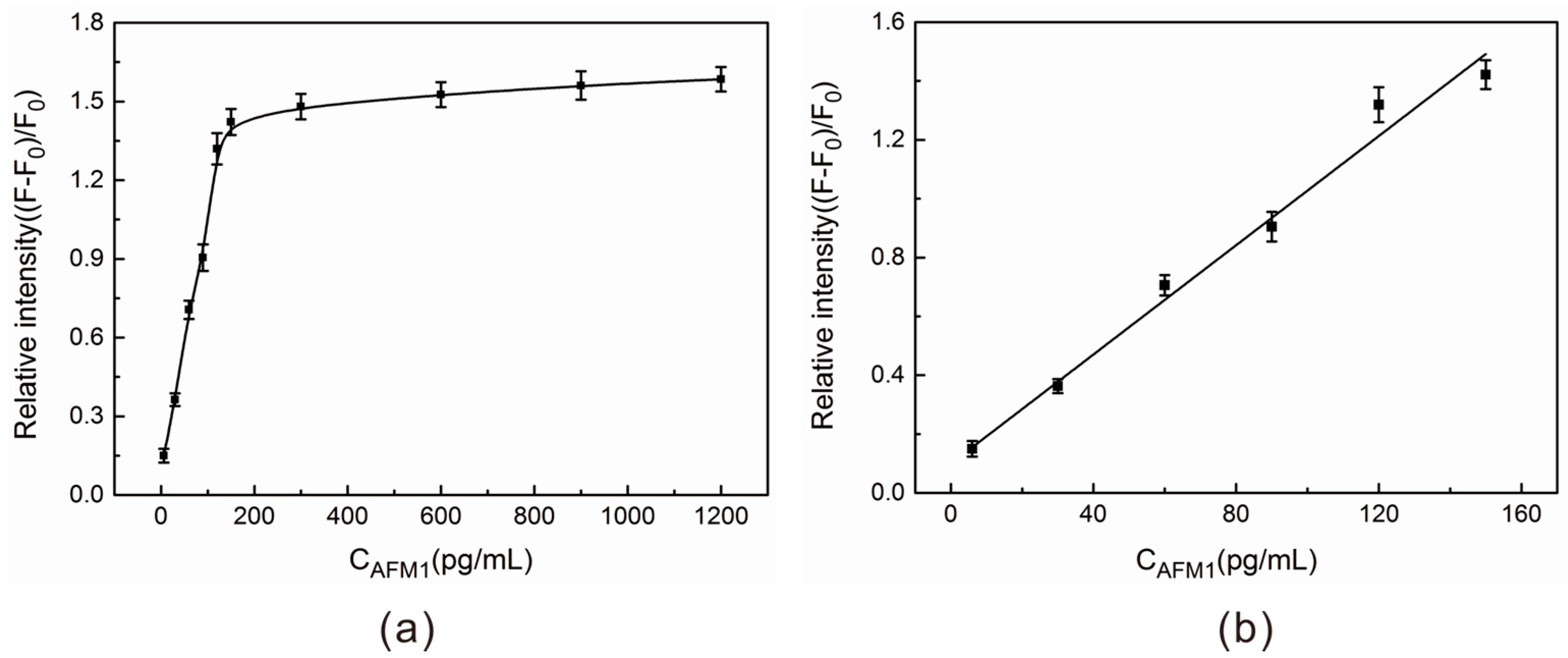
| Sample No. | Added (pg/mL) | Found (pg/mL) a | Recovery (%) | Relative Standard Deviation (RSD) (%) n = 3 |
|---|---|---|---|---|
| 1 | 10 | 9.20 | 92.0 | 3.4 |
| 2 | 70 | 65.64 | 93.8 | 4.8 |
| 3 | 120 | 127.85 | 106.5 | 4.9 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Yang, D.; Li, P.; Zhang, Q.; Zhang, W.; Ding, X.; Mao, J.; Wu, J. Palladium Nanoparticles-Based Fluorescence Resonance Energy Transfer Aptasensor for Highly Sensitive Detection of Aflatoxin M1 in Milk. Toxins 2017, 9, 318. https://doi.org/10.3390/toxins9100318
Li H, Yang D, Li P, Zhang Q, Zhang W, Ding X, Mao J, Wu J. Palladium Nanoparticles-Based Fluorescence Resonance Energy Transfer Aptasensor for Highly Sensitive Detection of Aflatoxin M1 in Milk. Toxins. 2017; 9(10):318. https://doi.org/10.3390/toxins9100318
Chicago/Turabian StyleLi, Hui, Daibin Yang, Peiwu Li, Qi Zhang, Wen Zhang, Xiaoxia Ding, Jin Mao, and Jing Wu. 2017. "Palladium Nanoparticles-Based Fluorescence Resonance Energy Transfer Aptasensor for Highly Sensitive Detection of Aflatoxin M1 in Milk" Toxins 9, no. 10: 318. https://doi.org/10.3390/toxins9100318






