Comparative Transcriptome Analysis of Penicillium citrinum Cultured with Different Carbon Sources Identifies Genes Involved in Citrinin Biosynthesis
Abstract
:1. Introduction
2. Results
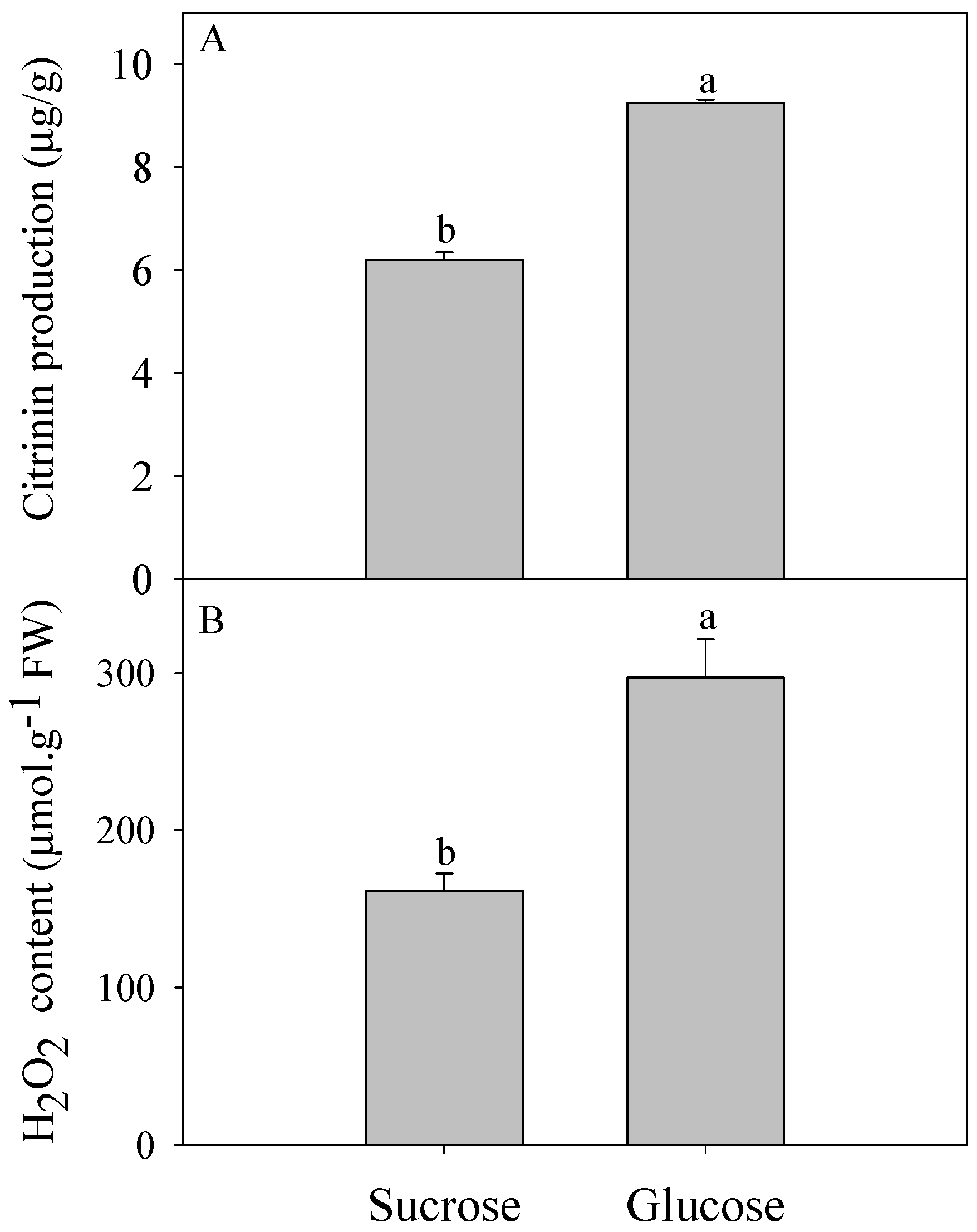
2.1. Effect of Different Carbon Sources on Citrinin Production by P. citrinum and Intracellular H2O2 Content
2.2. Sequencing, Assembly and Annotation
2.3. Analysis of DEGs between Sucrose- and Glucose-Cultured P. citrinum
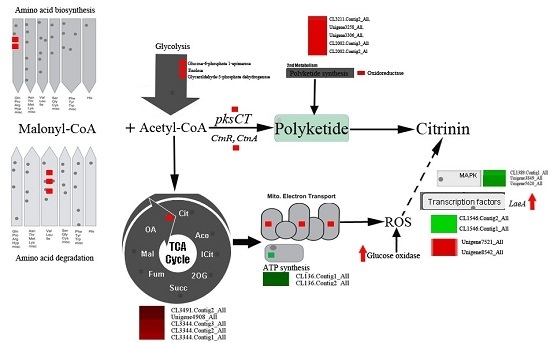
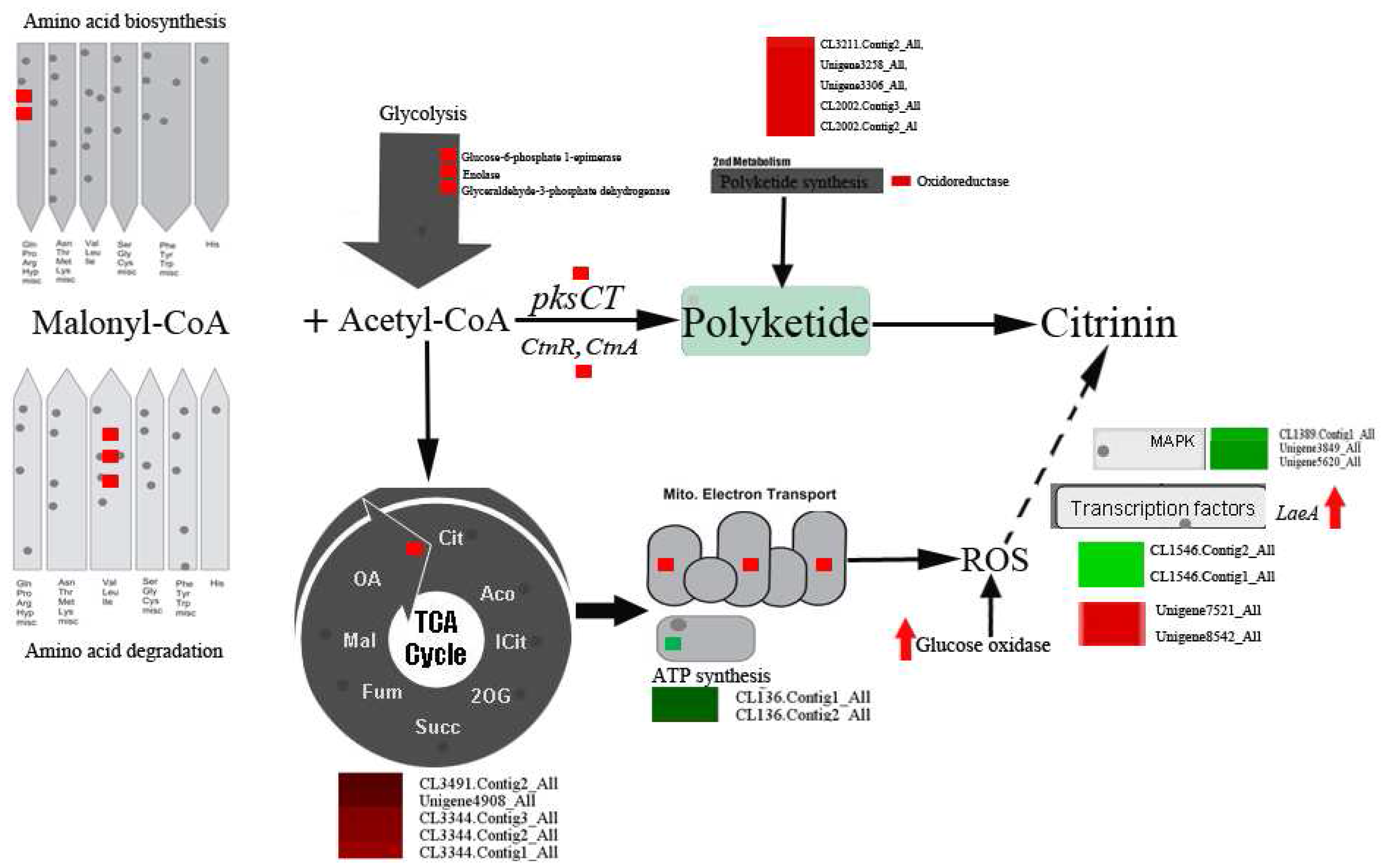
2.4. DEGs Involved in Secondary Metabolism in Sucrose- and Glucose-Cultured P. citrinum
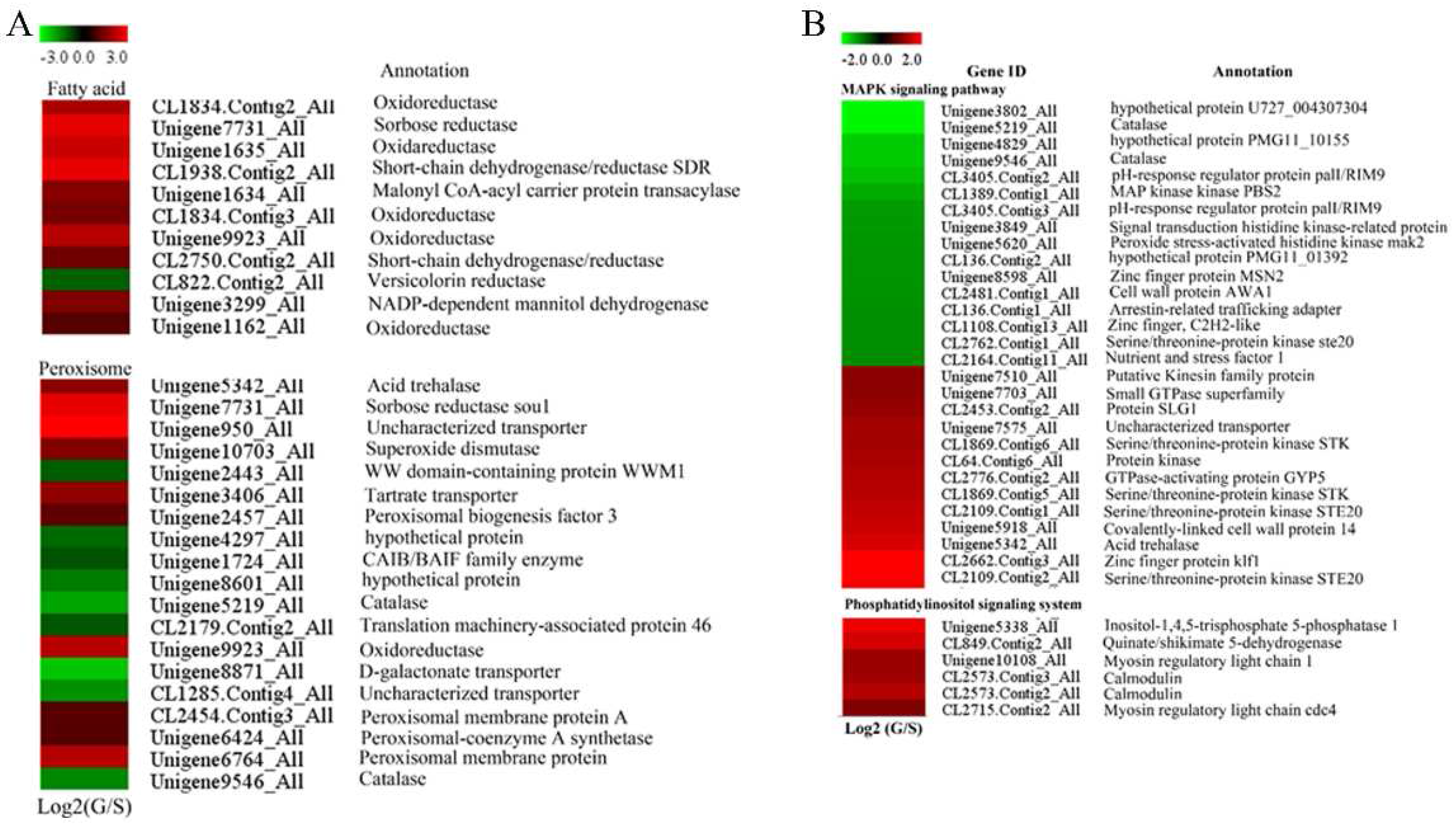
2.5. Peroxisome- and Proteasome-Related Genes
2.6. Genes Involved in Signal Transduction
2.7. Transcription Factors Genes
2.8. Genes Involved in Primary Metabolism
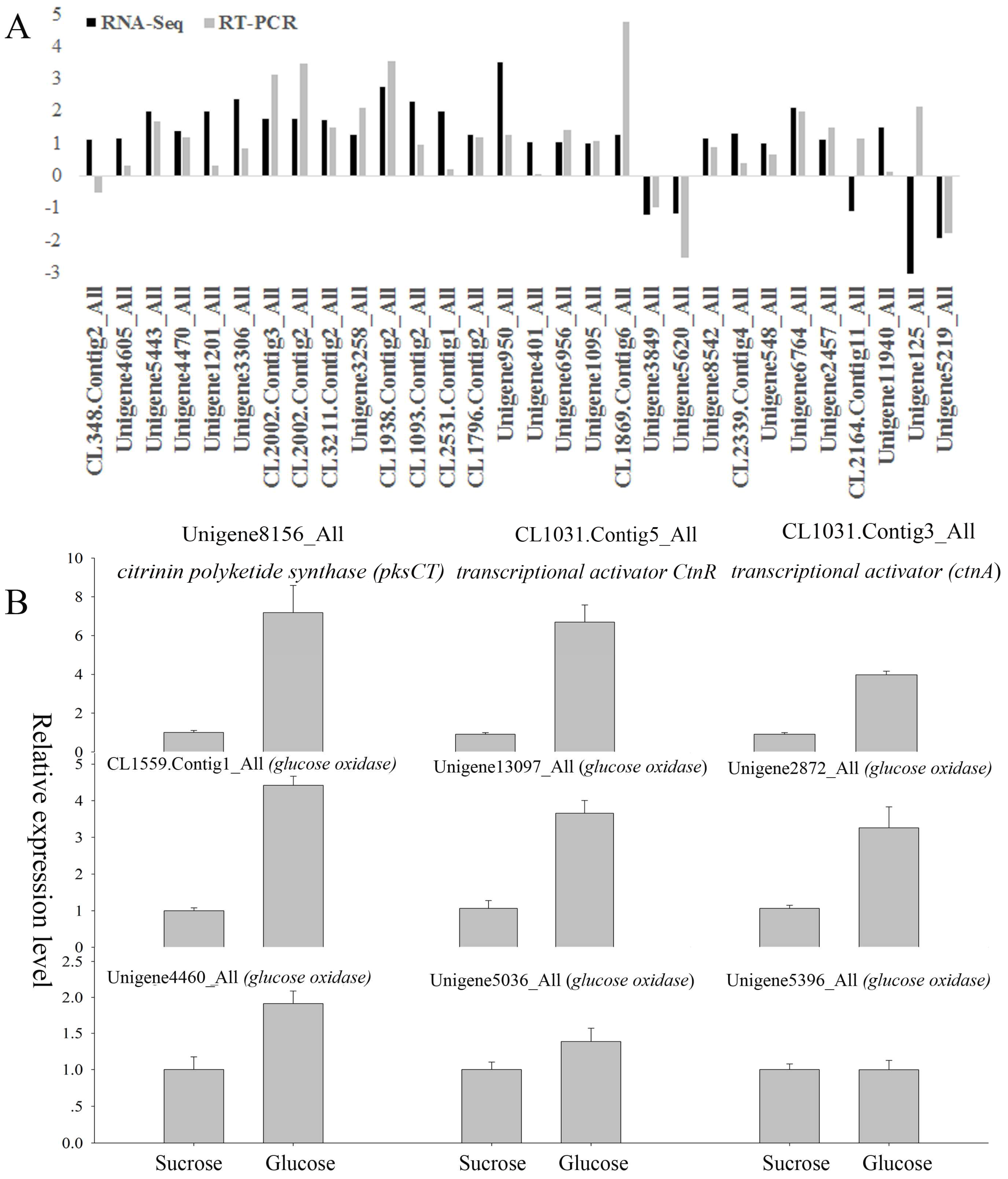
2.9. Quantitative Real-Time PCR (qRT-PCR) Confirmation
3. Discussions
3.1. Oxidative Stress Involved in Citrinin Biosynthesis by P. citrinum
3.2. DEGs Involved in Secondary Metabolism
3.3. DEGs Involved in Primary Metabolism
3.4. Signal Transduction and Transcription Factors (TFs)
4. Conclusions
5. Materials and Methods
5.1. Fungal Strain and Growth Condition
5.2. Citrinin Analysis
5.3. RNA Extraction, Library Construction and Sequencing
5.4. De Novo Assembly and Annotation of the P. citrinum Unigenes
5.5. DEGs between Different Samples and Function Enrichment
5.6. Evaluation of Genes Expression by qRT-PCR
5.7. Measurement of H2O2 Contents
5.8. Statistical Analyses
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Berthiller, F.; Crews, C.; Dall’Asta, C.; De Saeger, S.; Haesaert, G.; Karlovsky, P.; Oswald, I.P.; Seefelder, W.; Speijers, G.; Stroka, J. Masked mycotoxins: A review. Mol. Nutr. Food Res. 2013, 57, 165–186. [Google Scholar] [CrossRef] [PubMed]
- Comerio, R.; Pinto, V.E.F.; Vaamonde, G. Influence of water activity on Penicillium citrinum growth and kinetics of citrinin accumulation in wheat. Int. J. Food Microbiol. 1998, 42, 219–223. [Google Scholar] [CrossRef]
- Shimizu, T.; Kinoshita, H.; Ishihara, S.; Sakai, K.; Nagai, S.; Nihira, T. Polyketide synthase gene responsible for citrinin biosynthesis in Monascus purpureus. Appl. Environ. Microbiol. 2005, 71, 3453–3457. [Google Scholar] [CrossRef] [PubMed]
- Dutta, T.; Sahoo, R.; Sengupta, R.; Ray, S.S.; Bhattacharjee, A.; Ghosh, S. Novel cellulases from an extremophilic filamentous fungi Penicillium citrinum: Production and characterization. J. Ind. Microbiol. Biotechnol. 2008, 35, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, A.R.; Sheikhkarami, M.; Shokri, H.; Sabokbar, A. Genetic variability of citrinin-producing Penicillium Citrinum strains as occupational health hazards in Northern Iran. Arch. Ind. Hyg. Toxicol. 2012, 63, 489–496. [Google Scholar]
- Xu, B.J.; Jia, X.Q.; Gu, L.J.; Sung, C.K. Review on the qualitative and quantitative analysis of the mycotoxin citrinin. Food Control 2006, 17, 271–285. [Google Scholar] [CrossRef]
- Martins, M.L.; Gimeno, A.; Martins, H.M.; Bernardo, F. Co-occurrence of patulin and citrinin in Portuguese apples with rotten spots. Food Addit. Contam. 2002, 19, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Somorin, Y.; Akinyemi, A.; Bertuzzi, T.; Pietri, A. Co-occurrence of aflatoxins, ochratoxin A and citrinin in “egusi” melon (Colocynthis citrullus L.) seeds consumed in Ireland and the United Kingdom. Food Addit. Contam. B 2016, 9, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Aiko, V.; Mehta, A. Inhibitory Effect of Clove (Syzygium Aromaticum) on the Growth of Penicillium Citrinum and Citrinin Production. J. Food Saf. 2013, 33, 440–444. [Google Scholar] [CrossRef]
- Vrabcheva, T.; Usleber, E.; Dietrich, R.; Martlbauer, E. Co-occurrence of ochratoxin A and citrinin in cereals from Bulgarian villages with a history of Balkan endemic nephropathy. J. Agric. Food Chem. 2000, 48, 2483–2488. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.P.; Pan, Y.F.; Zou, L.H.; Xu, Y.; Huang, Z.B.; He, Q.H. Lower Citrinin Production by Gene Disruption of ctnB Involved in Citrinin Biosynthesis in Monascus aurantiacus Li AS3.4384. J. Agric. Food Chem. 2013, 61, 7397–7402. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Cox, R.J. The molecular steps of citrinin biosynthesis in fungi. Chem. Sci. 2016, 7, 2119–2127. [Google Scholar] [CrossRef]
- Sakai, K.; Kinoshita, H.; Shimizu, T.; Nihira, T. Construction of a citrinin gene cluster expression system in heterologous Aspergillus oryzae. J. Biosci. Bioeng. 2008, 106, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lam, C.W.; Tam, E.W.T.; Lee, K.C.; Yung, K.K.Y.; Leung, C.K.F.; Sze, K.H.; Lau, S.K.P.; Yuen, K.Y. The biosynthetic pathway for a thousand-year-old natural food colorant and citrinin in Penicillium marneffei. Sci. Rep. 2014, 4, 6728. [Google Scholar] [CrossRef] [PubMed]
- Hajjaj, H.; Klaebe, A.; Goma, G.; Blanc, P.J.; Barbier, E.; Francois, J. Medium-chain fatty acids affect citrinin production in the filamentous fungus Monascus ruber. Appl. Environ. Microbiol. 2000, 66, 1120–1125. [Google Scholar] [CrossRef] [PubMed]
- Hajjaj, H.; Francois, J.M.; Goma, G.; Blanc, P.J. Effect of amino acids on red pigments and citrinin production in Monascus ruber. J. Food Sci. 2012, 77, M156–M159. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.Y.; Hsu, W.H.; Shih, T.W.; Lin, C.H.; Pan, T.M. Proteomic insight into the effect of ethanol on citrinin biosynthesis pathway in Monascus putpureus NTU 568. Food Res. Int. 2014, 64, 733–742. [Google Scholar] [CrossRef]
- Mossini, S.A.G.; Kemmelmeier, C. Inhibition of citrinin production in Penicillium citrinum cultures by neem Azadirachta indica A. Juss (Meliaceae). Int. J. Mol. Sci. 2008, 9, 1676–1684. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.R.N.; Nurdijati, S.B.; Salleh, B. Efficacy of aqueous medicinal plant extracts on growth and citrinin production by Penicillium citrinum isolated from rice grains. Afr. J. Microbiol. Res. 2010, 4, 2562–2565. [Google Scholar]
- Noori, N.; Yahyaraeyat, R.; Khosravi, A.; Atefi, P.; Basti, A.A.; Akrami, F.; Bahonar, A.; Misaghi, A. Effect of Zataria Multiflora Boiss. essential oil on growth and citrinin production by Penicillium Citrinum in culture media and mozzarella Cheese. J. Food Saf. 2012, 32, 445–451. [Google Scholar] [CrossRef]
- Heperkan, D.; Dazkir, G.S.; Kansu, D.Z.; Guler, F.K. Influence of temperature on citrinin accumulation by Penicillium citrinum and Peniccillium verrucosum in black table olives. Toxin Rev. 2009, 28, 180–186. [Google Scholar] [CrossRef]
- Jiao, F.; Kawakami, A.; Nakajima, T. Effects of different carbon sources on trichothecene production and Tri gene expression by Fusarium graminearum in liquid culture. FEMS Microbiol. Lett. 2008, 285, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.Y.; Li, B.Q.; Tian, S.P. Effects of carbon, nitrogen and ambient pH on patulin production and related gene expression in Penicillium expansum. Int. J. Food Microbiol. 2015, 206, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Medina, A.; Mateo, E.M.; Valle-Algarra, F.M.; Mateo, F.; Mateo, R.; Jimenez, M. Influence of nitrogen and carbon sources on the production of ochratoxin A by ochratoxigenic strains of Aspergillus spp. isolated from grapes. Int. J. Food Microbiol. 2008, 122, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, L.M.; Lametsch, R.; Andersen, M.R.; Nielsen, P.V.; Frisvad, J.C. Proteome analysis of Aspergillus niger: Lactate added in starch-containing medium can increase production of the mycotoxin fumonisin B-2 by modifying acetyl-CoA metabolism. BMC Microbiol. 2009, 9, 255. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Barad, S.; Chen, Y.; Luo, X.; Tannous, J.; Dubey, A.; Glam Matana, N.; Tian, S.; Li, B.; Keller, N.; et al. LaeA regulation of secondary metabolism modulates virulence in Penicillium expansum and is mediated by sucrose. Mol. Plant Pathol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Barad, S.; Horowitz, S.B.; Kobiler, I.; Sherman, A.; Prusky, D. Accumulation of the mycotoxin patulin in the presence of gluconic acid contributes to pathogenicity of Penicillium expansum. Mol. Plant-Microbe Interact. 2014, 27, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, B.; Chávez, A.; Forero, A.; García-Huante, Y.; Romero, A.; Sánchez, M.; Rocha, D.; Sánchez, B.; Rodríguez-Sanoja, R.; Sánchez, S. Production of microbial secondary metabolites: Regulation by the carbon source. Crit. Rev. Microbiol. 2010, 36, 146–167. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, M.; Mateo, J.J.; Hinojo, M.J.; Mateo, R. Sugars and amino acids as factors affecting the synthesis of fumonisins in liquid cultures by isolates of the Gibberella fujikuroi complex. Int. J. Food Microbiol. 2003, 89, 185–193. [Google Scholar] [CrossRef]
- Sibthorp, C.; Wu, H.H.; Cowley, G.; Wong, P.W.H.; Palaima, P.; Morozov, I.Y.; Weedall, G.D.; Caddick, M.X. Transcriptome analysis of the filamentous fungus Aspergillus nidulans directed to the global identification of promoters. BMC Genom. 2013, 14, 847. [Google Scholar] [CrossRef] [PubMed]
- Meijueiro, M.L.; Santoyo, F.; Ramirez, L.; Pisabarro, A.G. Transcriptome characteristics of filamentous fungi deduced using high-throughput analytical technologies. Brief. Funct. Genom. 2014, 13, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Pasquali, M.; Lysoe, E.; Seong, K.; Menke, J.; Kistler, H. Transcriptome of Fusarium graminearum during plant infection and toxin biosynthesis. Phytopathology 2008, 98, S122. [Google Scholar]
- Montibus, M.; Pinson-Gadais, L.; Richard-Forget, F.; Barreau, C.; Ponts, N. Coupling of transcriptional response to oxidative stress and secondary metabolism regulation in filamentous fungi. Crit. Rev. Microbiol. 2015, 41, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Ponts, N.; Pinson-Gadais, L.; Barreau, C.; Richard-Forget, F.; Ouellet, T. Exogenous H2O2 and catalase treatments interfere with Tri genes expression in liquid cultures of Fusarium graminearum. FEBS Lett. 2007, 581, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Reverberi, M.; Punelli, F.; Scarpari, M.; Camera, E.; Zjalic, S.; Ricelli, A.; Fanelli, C.; Fabbri, A.A. Lipoperoxidation affects ochratoxin A biosynthesis in Aspergillus ochraceus and its interaction with wheat seeds. Appl. Microbiol. Biotechnol. 2010, 85, 1935–1946. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Heydt, M.; Stoll, D.; Schutz, P.; Geisen, R. Oxidative stress induces the biosynthesis of citrinin by Penicillium verrucosum at the expense of ochratoxin. Int. J. Food Microbiol. 2015, 192, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ugarte, N.; Petropoulos, I.; Friguet, B. Oxidized mitochondrial protein degradation and repair in aging and oxidative stress. Antioxid. Redox Signal. 2010, 13, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Delrio, L.A.; Sandalio, L.M.; Palma, J.M.; Bueno, P.; Corpas, F.J. Metabolism of oxygen radicals in peroxisomes and cellular implications. Free Radic. Biol. Med. 1992, 13, 557–580. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B.; del Rio, L.A. Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci. 2001, 6, 145–150. [Google Scholar] [CrossRef]
- Wong, C.M.; Wong, K.H.; Chen, X.D. Glucose oxidase: Natural occurrence, function, properties and industrial applications. Appl. Microbiol. Biotechnol. 2008, 78, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.Y.; Lee, H.Y.; Tang, Y.; Kim, C.Y.; Mathews, I.; Khosla, C. Structural and functional studies on SCO1815: A beta-ketoacyl-acyl carrier protein reductase from Streptomyces coelicolor A3(2). Biochemistry 2006, 45, 14085–14093. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Tsai, S.C. The type I fatty acid and polyketide synthases: A tale of two megasynthases. Nat. Prod. Rep. 2007, 24, 1041–1072. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Marti, E.; Zuzuarregui, A.; Gomar-Alba, M.; Gutierrez, D.; Gil, C.; del Olmo, M. Molecular response of Saccharomyces cerevisiae wine and laboratory strains to high sugar stress conditions. Int. J. Food Microbiol. 2011, 145, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.K.; Wright, P.C. The proteomic response of Saccharomyces cerevisiae in very high glucose conditions with amino acid supplementation. J. Proteome Res. 2008, 7, 4766–4774. [Google Scholar] [CrossRef] [PubMed]
- Barad, S.; Espeso, E.A.; Sherman, A.; Prusky, D. Ammonia activates pacC and patulin accumulation in an acidic environment during apple colonization by Penicillium expansum. Mol. Plant Pathol. 2016, 17, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Kohut, G.; Adam, A.L.; Fazekas, B.; Hornok, L. N-starvation stress induced FUM gene expression and fumonisin production is mediated via the HOG-type MAPK pathway in Fusarium proliferatum. Int. J. Food Microbiol. 2009, 130, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Ikner, A.; Shiozaki, K. Yeast signaling pathways in the oxidative stress response. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2005, 569, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Adam, A.L.; Kohut, G.; Hornok, L. Fphog1, a HOG-type MAP kinase gene, is involved in multistress response an Fusarium proliferatum. J. Basic Microbiol. 2008, 48, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Tomas-Cobos, L.; Casadome, L.; Mas, G.; Sanz, P.; Posas, F. Expression of the HXT1 low affinity glucose transporter requires the coordinated activities of the HOG and glucose signalling pathways. J. Biol. Chem. 2004, 279, 22010–22019. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, N.; Tokai, T.; Nishiuchi, T.; Takahashi-Ando, N.; Fujimura, M.; Kimura, M. Involvement of the osmosensor histidine kinase and osmotic stress-activated protein kinases in the regulation of secondary metabolism in Fusarium graminearum. Biochem. Biophys. Res. Commun. 2007, 363, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Stoll, D.; Schmidt-Heydt, M.; Geisen, R. Differences in the Regulation of Ochratoxin A by the HOG Pathway in Penicillium and Aspergillus in Response to High Osmolar Environments. Toxins 2013, 5, 1282–1298. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.P.; Xu, Y.; Huang, Z.B. Isolation and characterization of the citrinin biosynthetic gene cluster from Monascus aurantiacus. Biotechnol. Lett. 2012, 34, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Todd, R.B.; Andrianopoulos, A. Evolution of a fungal regulatory gene family: The Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol. 1997, 21, 388–405. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, G.; Kenmochi, Y.; Takano, Y.; Sweigard, J.; Farrall, L.; Furusawa, I.; Horino, O.; Kubo, Y. Novel fungal transcriptional activators, Cmr1p of Colletotrichum lagenarium and Pig1p of Magnaporthe grisea, contain Cys2His2 zinc finger and Zn(II)2Cys6 binuclear cluster DNA-binding motifs and regulate transcription of melanin biosynthesis genes in a developmentally specific manner. Mol. Microbiol. 2000, 38, 940–954. [Google Scholar] [PubMed]
- Flaherty, J.E.; Woloshuk, C.P. Regulation of fumonisin biosynthesis in Fusarium verticillioides by a zinc binuclear cluster-type gene, ZFR1. Appl. Environ. Microbiol. 2004, 70, 2653–2659. [Google Scholar] [CrossRef] [PubMed]
- Small, A.J.; Hynes, M.J.; Davis, M.A. The TamA protein fused to a DNA-binding domain can recruit AreA, the major nitrogen regulatory protein, to activate gene expression in Aspergillus nidulans. Genetics 1999, 153, 95–105. [Google Scholar] [PubMed]
- Baba, S.; Kinoshita, H.; Nihira, T. Identification and characterization of Penicillium citrinum VeA and LaeA as global regulators for ML-236B production. Curr. Genet. 2012, 58, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- TGICL 2.1. Available online: http://sourceforge.net/projects/tgicl/files/tgicl%20v2.1/ (accessed on 13 October 2016).
- BLASTx. Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 13 October 2016).
- Nr database. Available online: http://www.ncbi.nlm.nih.gov (accessed on 13 October 2016).
- Swiss-Prot protein database. Available online: http://www.expasy.ch/sprot (accessed on 13 October 2016).
- KEGG database. Available online: http://www.genome.jp/kegg (accessed on 13 October 2016).
- COG database. Available online: http://www.ncbi.nlm.nih.gov/COG (accessed on 13 October 2016).
- Blast2GO. Available online: http://www.blast2go.com/b2ghome (accessed on 13 October 2016).
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [PubMed]
- GO. Available online: http://www.geneontology.org (accessed on 17 October 2016).

| GO Term | Ontology * | Description | Number of Genes | Corrected-p-Value |
|---|---|---|---|---|
| GO: 0098656 | BP | anion transmembrane transport | 29 | 0.00010 |
| GO: 0003333 | BP | amino acid transmembrane transport | 25 | 0.00024 |
| GO: 0006734 | BP | NADH metabolic process | 6 | 0.0004 |
| GO: 0006116 | BP | NADH oxidation | 5 | 0.00179 |
| GO: 0006865 | BP | amino acid transport | 25 | 0.00198 |
| GO: 0071705 | BP | nitrogen compound transport | 40 | 0.00532 |
| GO: 0015862 | BP | uridine transport | 4 | 0.00619 |
| GO: 0015864 | BP | pyrimidine nucleoside transport | 4 | 0.00619 |
| GO: 0015849 | BP | organic acid transport | 27 | 0.00983 |
| GO: 0046942 | BP | carboxylic acid transport | 27 | 0.00983 |
| GO: 0055114 | BP | oxidation-reduction process | 115 | 0.02007 |
| GO: 0015711 | BP | organic anion transport | 29 | 0.02227 |
| GO: 0006072 | BP | glycerol-3-phosphate metabolic process | 6 | 0.03336 |
| Pathway ID | KEGG Pathway | Number of Genes | Q-Value |
|---|---|---|---|
| ko01110 | Biosynthesis of secondary metabolites | 123 | 0.003588954 |
| ko01100 | Metabolic pathways | 255 | 0.003588954 |
| ko00630 | Glyoxylate and dicarboxylate metabolism | 18 | 0.008545071 |
| ko00260 | Glycine, serine and threonine metabolism | 25 | 0.039994923 |
| ko00100 | Steroid biosynthesis | 17 | 0.039994923 |
| ko00061 | Fatty acid biosynthesis | 11 | 0.039994923 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, T.; Jiang, G.; Qu, H.; Wang, Y.; Xiong, Y.; Jian, Q.; Wu, Y.; Duan, X.; Zhu, X.; Hu, W.; et al. Comparative Transcriptome Analysis of Penicillium citrinum Cultured with Different Carbon Sources Identifies Genes Involved in Citrinin Biosynthesis. Toxins 2017, 9, 69. https://doi.org/10.3390/toxins9020069
Li T, Jiang G, Qu H, Wang Y, Xiong Y, Jian Q, Wu Y, Duan X, Zhu X, Hu W, et al. Comparative Transcriptome Analysis of Penicillium citrinum Cultured with Different Carbon Sources Identifies Genes Involved in Citrinin Biosynthesis. Toxins. 2017; 9(2):69. https://doi.org/10.3390/toxins9020069
Chicago/Turabian StyleLi, Taotao, Guoxiang Jiang, Hongxia Qu, Yong Wang, Yehui Xiong, Qijie Jian, Yu Wu, Xuewu Duan, Xiangrong Zhu, Wenzhong Hu, and et al. 2017. "Comparative Transcriptome Analysis of Penicillium citrinum Cultured with Different Carbon Sources Identifies Genes Involved in Citrinin Biosynthesis" Toxins 9, no. 2: 69. https://doi.org/10.3390/toxins9020069
APA StyleLi, T., Jiang, G., Qu, H., Wang, Y., Xiong, Y., Jian, Q., Wu, Y., Duan, X., Zhu, X., Hu, W., Wang, J., Gong, L., & Jiang, Y. (2017). Comparative Transcriptome Analysis of Penicillium citrinum Cultured with Different Carbon Sources Identifies Genes Involved in Citrinin Biosynthesis. Toxins, 9(2), 69. https://doi.org/10.3390/toxins9020069