A Fully Integrated In Vitro Diagnostic Microsystem for Pathogen Detection Developed Using a “3D Extensible” Microfluidic Design Paradigm
Abstract
1. Introduction
2. Materials and Methods
2.1. Design Paradigm of the “3D Extensible” Microfluidic Systems
2.2. Fabrication of “3D Extensible” Microfluidic Devices
2.3. Control and Detection Instrument
2.4. DNA Extraction and Loop-Mediated Isothermal Amplification (LAMP) Reaction
3. Results and Discussion
3.1. Unit Operations of the Microfluidic Platform
3.2. Design Process of a Fully Integrated System for Pathogen Detection
3.3. Operation of the iLAMP Microsystem
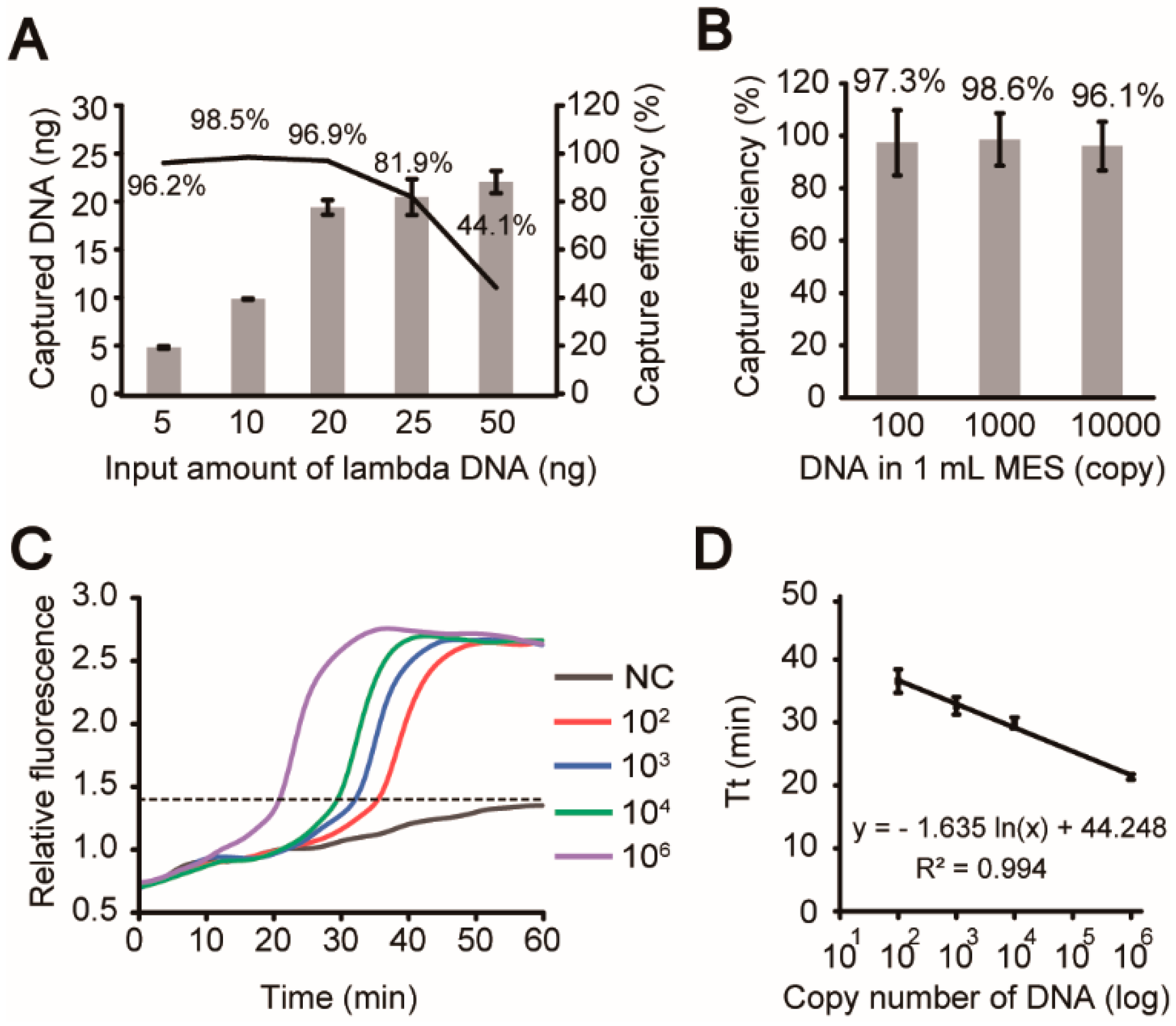
3.4. Evaluation of Analytical Steps
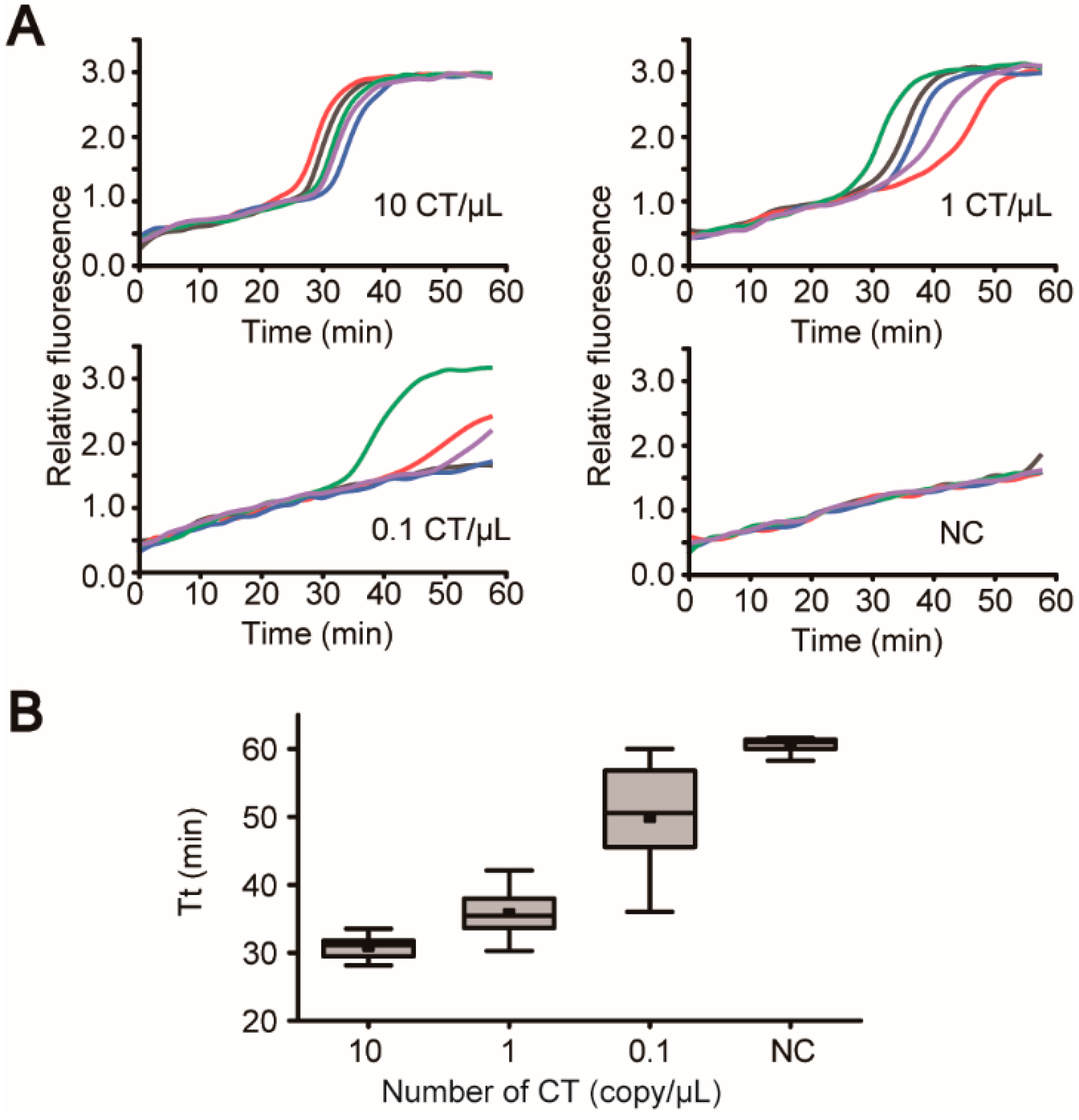
3.5. “Sample-In-Answer-Out” Analyses in the iLAMP System
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Yeo, L.Y.; Chang, H.C.; Chan, P.P.; Friend, J.R. Microfluidic devices for bioapplications. Small 2011, 7, 12–48. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.; Blumenfeld, N.R.; Laksanasopin, T.; Sia, S.K. Point-of-care diagnostics: Recent developments in a connected age. Anal. Chem. 2017, 89, 102–123. [Google Scholar] [CrossRef] [PubMed]
- Li, P. Microfluidics for IVD: In pursuit of the holy grail. J. Bioeng. Biomed. Sci. 2012, S8, e001. [Google Scholar]
- Chin, C.D.; Linder, V.; Sia, S.K. Commercialization of microfluidic point-of-care diagnostic devices. Lab Chip 2012, 12, 2118–2134. [Google Scholar] [CrossRef]
- Ahn, C.H.; Choi, J.W.; Beaucage, G.; Nevin, J.; Lee, J.B.; Puntambekar, A.; Lee, R.J.Y. Disposable smart lab on a chip for point-of-care clinical diagnostics. Proc. IEEE 2004, 92, 154–173. [Google Scholar] [CrossRef]
- Bhargava, K.C.; Thompson, B.; Malmstadt, N. Discrete elements for 3D microfluidics. Proc. Natl. Acad. Sci. USA 2014, 111, 15013–15018. [Google Scholar] [CrossRef]
- Haeberle, S.; Zengerle, R. Microfluidic platforms for lab-on-a-chip applications. Lab Chip 2007, 7, 1094–1110. [Google Scholar] [CrossRef]
- Mark, D.; Haeberle, S.; Roth, G.; von Stetten, F.; Zengerle, R. Microfluidic lab-on-a-chip platforms: requirements, characteristics and applications. Chem. Soc. Rev. 2010, 39, 1153–1182. [Google Scholar] [CrossRef]
- Strohmeier, O.; Keller, M.; Schwemmer, F.; Zehnle, S.; Mark, D.; von Stetten, F.; Zengerle, R.; Paust, N. Centrifugal microfluidic platforms: advanced unit operations and applications. Chem. Soc. Rev. 2015, 44, 6187–6229. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.W.; Phillips, S.T.; Whitesides, G.M.; Carrilho, E. Diagnostics for the developing world: Microfluidic paper-based analytical devices. Anal. Chem. 2010, 82, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Yetisen, A.K.; Akram, M.S.; Lowe, C.R. Paper-based microfluidic point-of-care diagnostic devices. Lab Chip 2013, 13, 2210–2251. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Chen, Z.; Cao, X.; Li, Z.; Stavrakis, S.; Choo, J.; deMello, A.J.; Howes, P.D.; He, N. A sample-in-digital-answer-out system for rapid detection and quantitation of infectious pathogens in bodily fluids. Anal. Bioanal. Chem. 2018, 410, 7019–7030. [Google Scholar] [CrossRef] [PubMed]
- Easley, C.J.; Karlinsey, J.M.; Bienvenue, J.M.; Legendre, L.A.; Roper, M.G.; Feldman, S.H.; Hughes, M.A.; Hewlett, E.L.; Merkel, T.J.; Ferrance, J.P.; et al. A fully integrated microfluidic genetic analysis system with sample-in-answer-out capability. Proc. Natl. Acad. Sci. USA 2006, 103, 19272–19277. [Google Scholar] [CrossRef] [PubMed]
- Gorkin, R.; Park, J.; Siegrist, J.; Amasia, M.; Lee, B.S.; Park, J.M.; Kim, J.; Kim, H.; Madou, M.; Cho, Y.K. Centrifugal microfluidics for biomedical applications. Lab Chip 2010, 10, 1758–1773. [Google Scholar] [CrossRef]
- Snyder, J.L.; Getpreecharsawas, J.; Fang, D.Z.; Gaborski, T.R.; Striemer, C.C.; Fauchet, P.M.; Borkholder, D.A.; McGrath, J.L. High-performance, low-voltage electroosmotic pumps with molecularly thin silicon nanomembranes. Proc. Natl. Acad. Sci. USA 2013, 110, 18425–18430. [Google Scholar] [CrossRef]
- Abdelgawad, M.; Wheeler, A.R. The digital revolution: A new paradigm for microfluidics. Adv. Mater. 2009, 21, 920–925. [Google Scholar] [CrossRef]
- Voiculescu, I.; Nordin, A.N. Acoustic wave based MEMS devices for biosensing applications. Biosens. Bioelectron. 2012, 33, 1–9. [Google Scholar] [CrossRef]
- Kim, J.; Johnson, M.; Hill, P.; Gale, B.K. Microfluidic sample preparation: Cell lysis and nucleic acid purification. Integr. Biol. 2009, 1, 574–586. [Google Scholar] [CrossRef]
- Parida, M.M.; Santhosh, S.R.; Dash, P.K.; Tripathi, N.K.; Saxena, P.; Ambuj, S.; Sahni, A.K.; Lakshmana-Rao, P.V.; Morita, K. Development and evaluation of reverse transcription-loop-mediated isothermal amplification assay for rapid and real-time detection of Japanese encephalitis virus. J. Clin. Microbiol. 2006, 44, 4172–4178. [Google Scholar] [CrossRef] [PubMed]
- Campos, C.D.M.; Gamage, S.S.T.; Jackson, J.M.; Witek, M.A.; Park, D.S.; Murphy, M.C.; Godwin, A.K.; Soper, S.A. Microfluidic-based solid phase extraction of cell free DNA. Lab Chip 2018, 18, 3459–3470. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Hsieh, T.M.; Lee, D.Y.; Ali, E.M.; Xie, H.; Looi, X.L.; Koay, E.S.; Li, M.H.; Ying, J.Y. A self-contained all-in-one cartridge for sample preparation and real-time PCR in rapid influenza diagnosis. Lab Chip 2010, 10, 3103–3111. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.V.; Nguyen, V.D.; Lee, E.Y.; Seo, T.S. Point-of-care genetic analysis for multiplex pathogenic bacteria on a fully integrated centrifugal microdevice with a large-volume sample. Biosens. Bioelectron. 2019, 136, 132–139. [Google Scholar] [CrossRef]
- Hoffmann, J.; Mark, D.; Lutz, S.; Zengerle, R.; von Stetten, F. Pre-storage of liquid reagents in glass ampoules for DNA extraction on a fully integrated lab-on-a-chip cartridge. Lab Chip 2010, 10, 1480–1484. [Google Scholar] [CrossRef]
- Ferguson, B.S.; Buchsbaum, S.F.; Wu, T.T.; Hsieh, K.; Xiao, Y.; Sun, R.; Soh, H.T. Genetic analysis of H1N1 influenza virus from throat swab samples in a microfluidic system for point-of-care diagnostics. J. Am. Chem. Soc. 2011, 133, 9129–9135. [Google Scholar] [CrossRef]
- Sun, Y.; Haglund, T.A.; Rogers, A.J.; Ghanim, A.F.; Sethu, P. Review: Microfluidics technologies for blood-based cancer liquid biopsies. Anal. Chim. Acta 2018, 1012, 10–29. [Google Scholar] [CrossRef]
- Culbertson, C.T.; Mickleburgh, T.G.; Stewart-James, S.A.; Sellens, K.A.; Pressnall, M. Micro total analysis systems: Fundamental advances and biological applications. Anal. Chem. 2014, 86, 95–118. [Google Scholar] [CrossRef]
- Hsieh, Y.-F.; Yang, A.-S.; Chen, J.-W.; Liao, S.-K.; Su, T.-W.; Yeh, S.-H.; Chen, P.-J.; Chen, P.-H. A Lego®-like swappable fluidic module for bio-chem applications. Sens. Actuators B 2014, 204, 489–496. [Google Scholar] [CrossRef]
- Vittayarukskul, K.; Lee, A.P. A truly Lego (R)-like modular microfluidics platform. J. Micromech. Microeng. 2017, 27, 035004. [Google Scholar] [CrossRef]
- Meng, Z.-J.; Wang, W.; Liang, X.; Zheng, W.-C.; Deng, N.-N.; Xie, R.; Ju, X.-J.; Liu, Z.; Chu, L.-Y. Plug-n-play microfluidic systems from flexible assembly of glass-based flow-control modules. Lab Chip 2015, 15, 1869–1878. [Google Scholar] [CrossRef] [PubMed]
- Yuen, P.K. A reconfigurable stick-n-play modular microfluidic system using magnetic interconnects. Lab Chip 2016, 16, 3700–3707. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Lin, S.; Wang, C.; Hu, J.; Li, C.; Zhuang, Z.; Zhou, Y.; Mathies, R.A.; Yang, C.J. PMMA/PDMS valves and pumps for disposable microfluidics. Lab Chip 2009, 9, 3088–3094. [Google Scholar] [CrossRef] [PubMed]
- Ogilvie, I.R.; Sieben, V.J.; Cortese, B.; Mowlem, M.C.; Morgan, H. Chemically resistant microfluidic valves from Viton® membranes bonded to COC and PMMA. Lab Chip 2011, 11, 2455–2459. [Google Scholar] [CrossRef] [PubMed]
- Gan, W.; Gu, Y.; Han, J.; Li, C.X.; Sun, J.; Liu, P. Chitosan-modified filter paper for nucleic acid extraction and “in situ PCR” on a thermoplastic microchip. Anal. Chem. 2017, 89, 3568–3575. [Google Scholar] [CrossRef] [PubMed]
- Meyer, T. Diagnostic procedures to detect Chlamydia trachomatis infections. Microorganisms 2016, 4, 25. [Google Scholar] [CrossRef]
- Hui, J.; Gu, Y.; Zhu, Y.; Chen, Y.; Guo, S.J.; Tao, S.C.; Zhang, Y.; Liu, P. Multiplex sample-to-answer detection of bacteria using a pipette-actuated capillary array comb with integrated DNA extraction, isothermal amplification, and smartphone detection. Lab Chip 2018, 18, 2854–2864. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geng, Z.; Gu, Y.; Li, S.; Lin, B.; Liu, P. A Fully Integrated In Vitro Diagnostic Microsystem for Pathogen Detection Developed Using a “3D Extensible” Microfluidic Design Paradigm. Micromachines 2019, 10, 873. https://doi.org/10.3390/mi10120873
Geng Z, Gu Y, Li S, Lin B, Liu P. A Fully Integrated In Vitro Diagnostic Microsystem for Pathogen Detection Developed Using a “3D Extensible” Microfluidic Design Paradigm. Micromachines. 2019; 10(12):873. https://doi.org/10.3390/mi10120873
Chicago/Turabian StyleGeng, Zhi, Yin Gu, Shanglin Li, Baobao Lin, and Peng Liu. 2019. "A Fully Integrated In Vitro Diagnostic Microsystem for Pathogen Detection Developed Using a “3D Extensible” Microfluidic Design Paradigm" Micromachines 10, no. 12: 873. https://doi.org/10.3390/mi10120873
APA StyleGeng, Z., Gu, Y., Li, S., Lin, B., & Liu, P. (2019). A Fully Integrated In Vitro Diagnostic Microsystem for Pathogen Detection Developed Using a “3D Extensible” Microfluidic Design Paradigm. Micromachines, 10(12), 873. https://doi.org/10.3390/mi10120873




