Quantification and Imaging of Antigens on Cell Surface with Lipid-Encapsulated Fluorescent Nanodiamonds
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
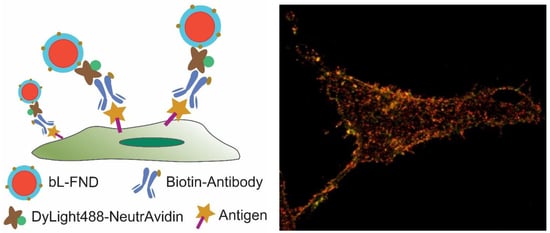
3.1. Flow Cytometry and Confocal Fluorescence Imaging of Cell Surface Antigens
3.2. Quantification of Cell Surface Antigens by Magnetic Modulated Fluorescence
4. Conclusions and Future Work
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Takahashi, T.; Shiku, H. Cell surface antigens: Invaluable landmarks reflecting the nature of cells. Cancer Immun. 2012, 12, 2. [Google Scholar]
- Várady, G.; Cserepes, J.; Németh, A.; Szabó, E.; Sarkadi, B. Cell surface membrane proteins as personalized biomarkers: Where we stand and where we are headed. Biomark. Med. 2013, 7, 803–819. [Google Scholar] [CrossRef]
- Belov, L.; Zhou, J.; Christopherson, R.I. Cell surface markers in colorectal cancer prognosis. Int. J. Mol. Sci. 2010, 12, 78–113. [Google Scholar] [CrossRef]
- Dean, L. Blood group antigens are surface markers on the red blood cell membrane. In Blood Groups and Red Cell Antigens; Laura, D., Ed.; National Center for Biotechnology Information: Bethesda, MD, USA, 2005. [Google Scholar]
- Delves, P.J. CD antigens. In eLS; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2016; pp. 1–26. [Google Scholar] [CrossRef]
- Grimm, D.; Bauer, J.; Pietsch, J.; Infanger, M.; Eucker, J.; Eilles, C.; Schoenberger, J. Diagnostic and therapeutic use of membrane proteins in cancer cells. Curr. Med. Chem. 2011, 18, 176–190. [Google Scholar] [CrossRef]
- Bishop, G.A.; Hwang, J. Use of a cellular Elisa for the detection of cell-surface antigens. BioTechniques 1992, 12, 326–330. [Google Scholar] [PubMed]
- Ravindranath, M.H.; Bauer, P.M.; Cornillez-Ty, C.; Garcia, J.; Morton, D.L. Quantitation of the density of cell surface carbohydrate antigens on cancer cells with a sensitive cell-suspension ELISA. J. Immunol. Methods 1996, 197, 51–67. [Google Scholar] [CrossRef]
- Lourenco, E.V.; Roque-Barreira, M.C. Immunoenzymatic quantitative analysis of antigens expressed on the cell surface (cell-ELISA). Methods Mol. Biol. 2010, 588, 301–309. [Google Scholar] [CrossRef]
- Kim, H.; Tsuruta, S.; Arakawa, H.; Osada, T.; Ikai, A. Quantitative analysis of the number of antigens immobilized on a glass surface by AFM. Ultramicroscopy 2004, 100, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Tijssen, P. Practice and Theory of Enzyme Immunoassays; Elsevier: Amsterdam, The Netherlands, 1985; Volume 15. [Google Scholar]
- de la Rica, R.; Stevens, M.M. Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nat. Nanotechnol. 2012, 7, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.P.; Cui, Y.L.; Chen, G.A. Nanoparticle-based immunoassays in the biomedical field. Analyst 2013, 138, 981–990. [Google Scholar] [CrossRef]
- Roth, K.A.; Brenner, J.W.; Selznick, L.A.; Gokden, M.; Lorenz, R.G. Enzyme-based antigen localization and quantitation in cell and tissue samples (Midwestern assay). J. Histochem. Cytochem. 1997, 45, 1629–1641. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.J.; Kang, M.W.; Chang, H.C.; Chen, K.M.; Yu, Y.C. Bright fluorescent nanodiamonds: No photobleaching and low cytotoxicity. J. Am. Chem. Soc. 2005, 127, 17604–17605. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.-C.; Lee, H.-Y.; Chen, K.; Lim, T.-S.; Wu, H.-Y.; Lin, P.-K.; Wei, P.-K.; Tsao, P.-H.; Chang, H.-C.; Fann, W. Characterization and application of single fluorescent nanodiamonds as cellular biomarkers. Proc. Natl. Acad. Sci. USA 2007, 104, 727–732. [Google Scholar] [CrossRef] [Green Version]
- Chapman, R.; Plakhotnik, T. Background-free imaging of luminescent nanodiamonds using external magnetic field for contrast enhancement. Opt. Lett. 2013, 38, 1847–1849. [Google Scholar] [CrossRef]
- Kuo, Y.; Hsu, T.-Y.; Wu, Y.C.; Hsu, J.H.; Chang, H.C. Fluorescence lifetime imaging microscopy of nanodiamonds in vivo. Proc. SPIE 2013, 8635, 863503–863508. [Google Scholar] [CrossRef]
- Kuo, Y.; Hsu, T.Y.; Wu, Y.C.; Chang, H.C. Fluorescent nanodiamond as a probe for the intercellular transport of proteins in vivo. Biomaterials 2013, 34, 8352–8360. [Google Scholar] [CrossRef]
- Wu, T.J.; Tzeng, Y.K.; Chang, W.W.; Cheng, C.A.; Kuo, Y.; Chien, C.H.; Chang, H.C.; Yu, J. Tracking the engraftment and regenerative capabilities of transplanted lung stem cells using fluorescent nanodiamonds. Nat. Nanotechnol. 2013, 8, 682–689. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.K.; Bumb, A.; Wu, X.; Sochacki, K.A.; Kellman, P.; Brechbiel, M.W.; Neuman, K.C. Wide-field in vivo background free imaging by selective magnetic modulation of nanodiamond fluorescence. Biomed. Opt. Express 2014, 5, 1190–1202. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, F.J.; Chen, Y.W.; Huang, Y.K.; Lee, H.M.; Lin, C.H.; Chang, H.C. Correlative light-electron microscopy of lipid-encapsulated fluorescent nanodiamonds for nanometric localization of cell surface antigens. Anal. Chem. 2018, 90, 1566–1571. [Google Scholar] [CrossRef]
- Prabhakar, N.; Peurla, M.; Koho, S.; Deguchi, T.; Näreoja, T.; Chang, H.C.; Rosenholm, J.M.; Hänninen, P.E. STED-TEM correlative microscopy leveraging nanodiamonds as intracellular dual-contrast markers. Small 2018, 14, 1701807. [Google Scholar] [CrossRef]
- Su, L.J.; Wu, M.S.; Hui, Y.Y.; Chang, B.M.; Pan, L.; Hsu, P.C.; Chen, Y.T.; Ho, H.N.; Huang, Y.H.; Ling, T.Y.; Hsu, H.H.; Chang, H.C. Fluorescent nanodiamonds enable quantitative tracking of human mesenchymal stem cells in miniature pigs. Sci. Rep. 2017, 7, 45607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sotoma, S.; Hsieh, F.J.; Chen, Y.W.; Tsai, P.C.; Chang, H.C. Highly stable lipid-encapsulation of fluorescent nanodiamonds for bioimaging applications. Chem. Commun. (Camb) 2018, 54, 1000–1003. [Google Scholar] [CrossRef]
- Su, L.J.; Fang, C.Y.; Chang, Y.T.; Chen, K.M.; Yu, Y.C.; Hsu, J.H.; Chang, H.C. Creation of high density ensembles of nitrogen-vacancy centers in nitrogen-rich type Ib nanodiamonds. Nanotechnology 2013, 24, 315702. [Google Scholar] [CrossRef]
- Chang, B.M.; Lin, H.H.; Su, L.J.; Lin, W.D.; Lin, R.J.; Tzeng, Y.K.; Lee, R.T.; Lee, Y.C.; Yu, A.L.; Chang, H.C. Highly fluorescent nanodiamonds protein-functionalized for cell labeling and targeting. Adv. Funct. Mater. 2013, 23, 5737–5745. [Google Scholar] [CrossRef]
- Davis, K.A.; Abrams, B.; Iyer, S.B.; Hoffman, R.A.; Bishop, J.E. Determination of CD4 antigen density on cells: Role of antibody valency, avidity, clones, and conjugation. Cytometry 1998, 33, 197–205. [Google Scholar] [CrossRef] [Green Version]
- Pannu, K.K.; Joe, E.T.; Iyer, S.B. Performance evaluation of QuantiBRITE phycoerythrin beads. Cytometry 2001, 45, 250–258. [Google Scholar] [CrossRef]
- Angelova, B.; Angelova, A. Nanoscale clustering of the neurotrophin receptor TrkB revealed by super-resolution STED microscopy. Nanoscale 2017, 9, 9797–9804. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Camejo, S.; Adam, M.P.; Besbes, M.; Hugonin, J.P.; Jacques, V.; Greffet, J.J.; Roch, J.F.; Hell, S.W.; Treussart, F. Stimulated emission depletion microscopy resolves individual nitrogen vacancy centers in diamond nanocrystals. ACS Nano 2013, 7, 10912–10919. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsieh, F.-J.; Chen, Y.-W.; Hui, Y.Y.; Lin, C.-H.; Chang, H.-C. Quantification and Imaging of Antigens on Cell Surface with Lipid-Encapsulated Fluorescent Nanodiamonds. Micromachines 2019, 10, 304. https://doi.org/10.3390/mi10050304
Hsieh F-J, Chen Y-W, Hui YY, Lin C-H, Chang H-C. Quantification and Imaging of Antigens on Cell Surface with Lipid-Encapsulated Fluorescent Nanodiamonds. Micromachines. 2019; 10(5):304. https://doi.org/10.3390/mi10050304
Chicago/Turabian StyleHsieh, Feng-Jen, Yen-Wei Chen, Yuen Yung Hui, Chun-Hung Lin, and Huan-Cheng Chang. 2019. "Quantification and Imaging of Antigens on Cell Surface with Lipid-Encapsulated Fluorescent Nanodiamonds" Micromachines 10, no. 5: 304. https://doi.org/10.3390/mi10050304