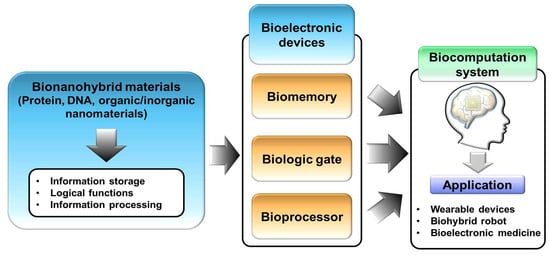
Development of Bioelectronic Devices Using Bionanohybrid Materials for Biocomputation System
Abstract
:1. Introduction
2. Biomemory
2.1. Multilevel Biomemory Devices
2.2. Electrochemical Signal-Enhanced Biomemory Device
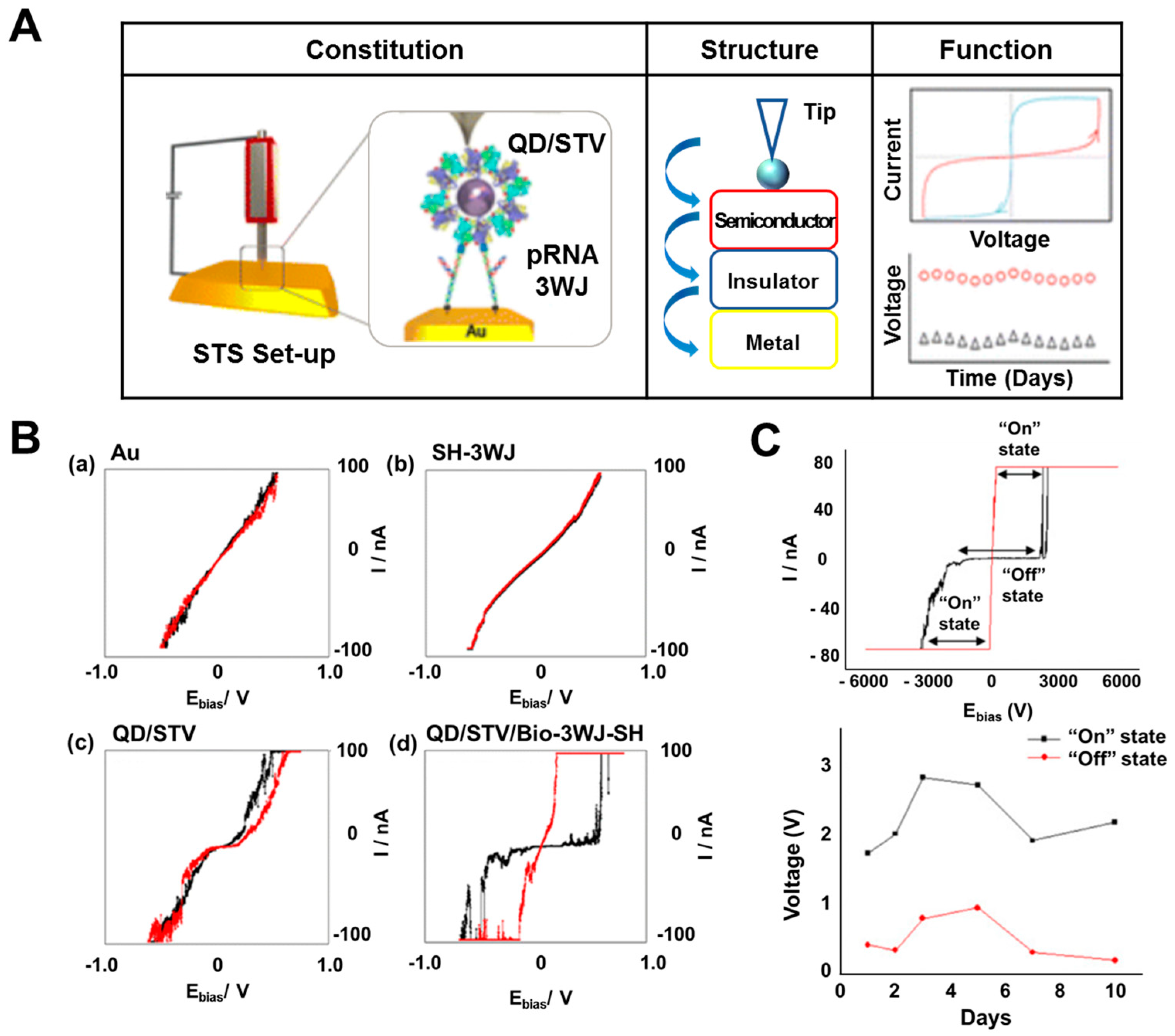
2.3. Resistive Biomemory Device
3. Biologic Gate
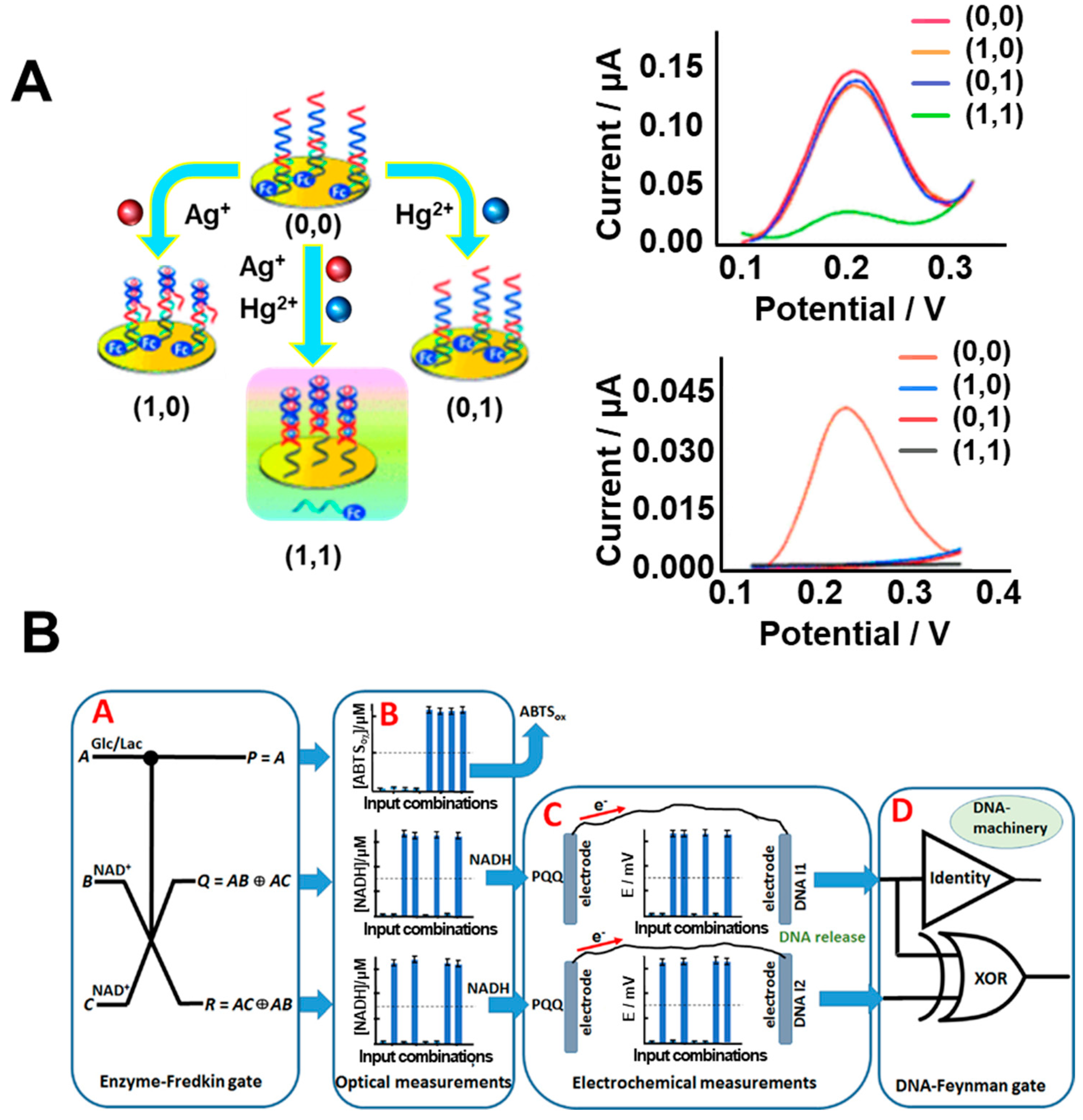
3.1. DNA-Based Biologic Gate
3.2. Protein/DNA-Based Biologic Gate
3.3. Analog Decision Mimicking Bioelectronic Device
4. Bioprocessor
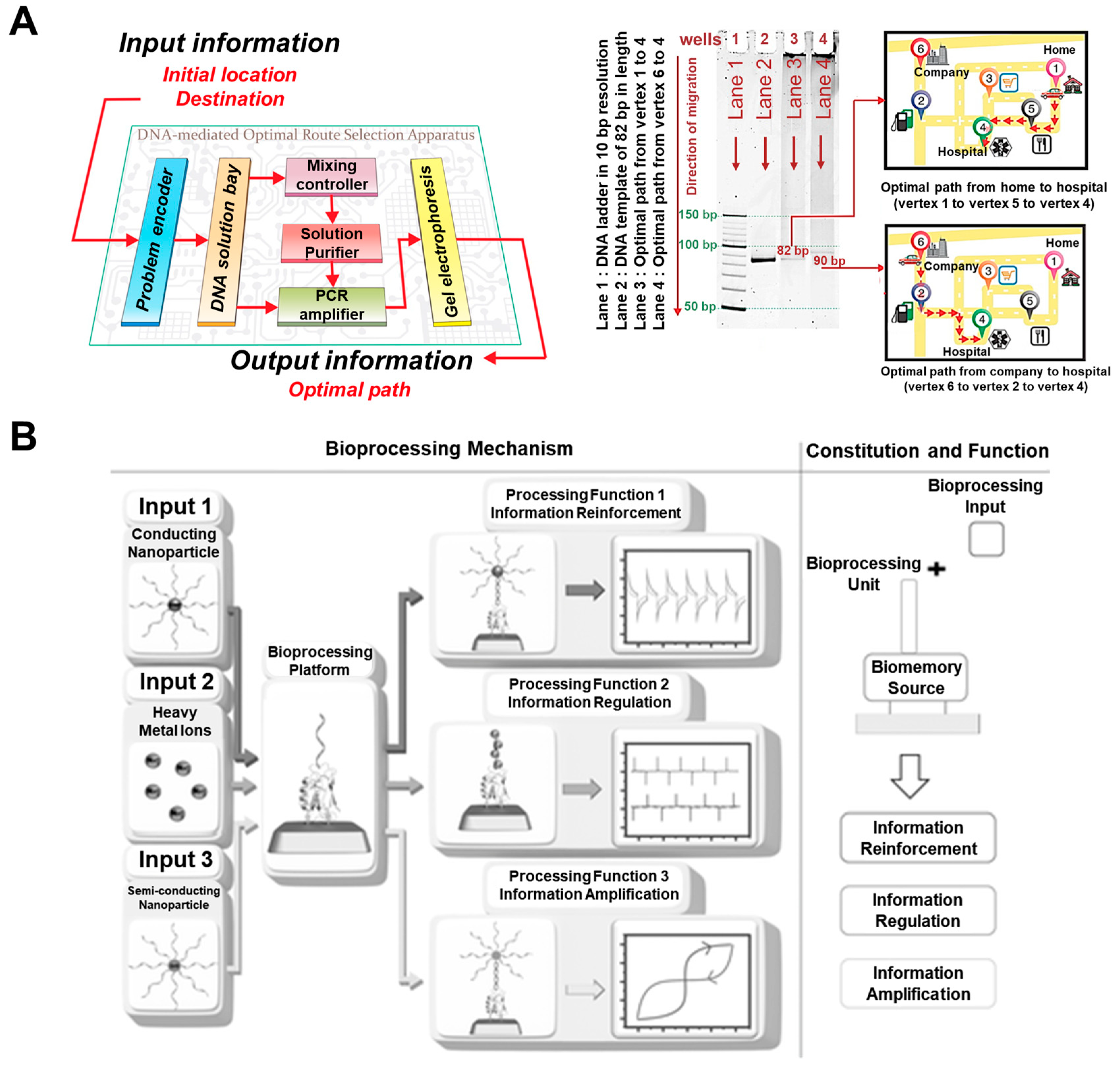
4.1. DNA-Based Bioprocessor
4.2. Bioprocessor Based on Bionanohybrid Material
5. Future Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Willner, I.; Willner, B. Biomaterials integrated with electronic elements: En route to bioelectronics. Trends Biotechnol. 2001, 19, 222–230. [Google Scholar] [CrossRef]
- Goode, J.A.; Rushworth, J.V.H.; Millner, P.A. Biosensor Regeneration: A Review of Common Techniques and Outcomes. Langmuir 2015, 31, 6267–6276. [Google Scholar] [CrossRef]
- Lee, T.; Lee, Y.; Park, S.Y.; Hong, K.; Kim, Y.; Park, C.; Chung, Y.-H.; Lee, M.-H.; Min, J. Fabrication of Electrochemical Biosensor Composed of Multi-functional DNA Structure/Au Nanospike on Micro-gap/PCB System for Detecting Troponin I in Human Serum. Colloids Surf. B 2019, 175, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Park, S.Y.; Jang, H.; Kim, G.H.; Lee, Y.; Park, C.; Mohammadniaei, M.; Lee, M.-H.; Min, J. Fabrication of Electrochemical Biosensor consisted of Multi-functional DNA Structure/porous Au Nanoparticle for Avian Influenza Virus (H5N1) in Human Serum. Mater. Sci. Eng. C 2019, 99, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.A.; Barker, L.; Semprini, L.; Minot, E.D. Graphene Biotransistor Interfaced with a Nitrifying Biofilm. Environ. Sci. Technol. Lett. 2015, 2, 118–122. [Google Scholar] [CrossRef]
- Gorton, L.; Lindgren, A.; Larsson, T.; Munteanu, F.D.; Ruzgas, T.; Gazaryan, I. Direct electron transfer between heme-containing enzymes and electrodes as basis for third generation biosensors. Anal. Chim. Acta 1999, 400, 91–108. [Google Scholar] [CrossRef]
- Akkilic, N.; Kamran, M.; Stan, R.; Sanghamitra, N.J.M. Voltage-controlled fluorescence switching of a single redox protein. Biosens. Bioelectron. 2015, 67, 747–751. [Google Scholar] [CrossRef]
- Macchia, E.; Alberga, D.; Manoli, K.; Mangiatordi, G.F.; Magliulo, M.; Palazzo, G.; Giordano, F.; Lattanzi, G.; Torsi, L. Organic bioelectronics probing conformational changes in surface confined proteins. Sci. Rep. 2016, 6, 28085. [Google Scholar] [CrossRef] [Green Version]
- Berggren, M.; Richter-Dahlfors, A. Organic Bioelectronics. Adv. Mater. 2007, 19, 3201–3213. [Google Scholar] [CrossRef]
- Rivnay, J.; Owens, R.M.; Malliaras, G.G. The Rise of Organic Bioelectronics. Chem. Mater. 2014, 26, 679–685. [Google Scholar] [CrossRef]
- Zhang, A.; Lieber, C.M. Nano-Bioelectronics. Chem. Rev. 2016, 116, 215–257. [Google Scholar] [CrossRef]
- Bostick, C.D.; Mukhopadhyay, S.; Pecht, I. Protein bioelectronics: A review of what we do and do not know. Rep. Prog. Phys. 2018, 81, 026601. [Google Scholar] [CrossRef]
- Kim, M.J.; Yang, M.-S.; Kwon, H.T.; Song, J.M. Low noise bipolar photodiode array protein chip based on on-chip bioassay for the detection of E. coli O157:H7. Biomed. Microdevices 2007, 9, 565–572. [Google Scholar] [CrossRef]
- Vistas, C.R.; Soares, S.S.; Rodrigues, R.M.M.; Chu, V.; Conde, J.P.; Ferreira, G.N.M. An amorphous silicon photodiode microfluidic chip to detect nanomolar quantities of HIV-1 virion infectivity factor. Analyst 2014, 139, 3709–3713. [Google Scholar] [CrossRef]
- Ingebrandt, S. Bioelectronics: Sensing beyond the limit. Nat. Nanotechnol. 2015, 10, 734–735. [Google Scholar] [CrossRef]
- Thudi, L.; Jasti, L.S.; Swarnalatha, Y.; Fadnavis, N.W.; Mulani, K.; Deokar, S.; Ponrathnam, S. Adsorption induced enzyme denaturation: The role of protein surface in adsorption induced protein denaturation on allyl glycidyl ether (AGE)-ethylene glycol dimethacrylate (EGDM) copolymers. Colloids Surf. B 2012, 90, 184–190. [Google Scholar] [CrossRef]
- Lee, T.; Yoo, S.-Y.; Chung, Y.-H.; Min, J.; Choi, J.-W. Signal Enhancement of Electrochemical Biomemory Device Composed of Recombinant Azurin/Gold Nanoparticle. Electroanalysis 2011, 23, 2023–2029. [Google Scholar] [CrossRef]
- Yoon, J.; Lee, T.; Bapurao, G.B.; Jo, J.; Oh, B.-K.; Choi, J.-W. Electrochemical H2O2 biosensor composed of myoglobin on MoS2 nanoparticle-graphene oxide hybrid structure. Biosens. Bioelectron. 2017, 93, 14–20. [Google Scholar] [CrossRef]
- Zebda, A.; Gondran, C.; Goff, A.L.; Holzinger, M.; Cinquin, P.; Cosnier, S. Mediatorless high-power glucose biofuel cells based on compressed carbon nanotube-enzyme electrodes. Nat. Commun. 2011, 2, 370. [Google Scholar] [CrossRef] [Green Version]
- Zuo, X.; He, S.; Li, D.; Peng, C.; Huang, Q.; Song, S.; Fan, C. Graphene Oxide-Facilitated Electron Transfer of Metalloproteins at Electrode Surfaces. Langmuir 2010, 26, 1936–1939. [Google Scholar] [CrossRef]
- Kwak, S.K.; Lee, G.S.; Ahn, D.J.; Choi, J.-W. Pattern formation of cytochrome c by microcontact printing and dip-pen nanolithography. Mater. Sci. Eng. C 2004, 24, 151–155. [Google Scholar] [CrossRef]
- Wu, X.; Xiao, T.; Luo, Z.; He, R.; Cao, Y.; Guo, Z.; Zhang, W.; Chen, Y. A micro-/nano-chip and quantum dots-based 3D cytosensor for quantitative analysis of circulating tumor cells. J. Nanobiotechnol. 2018, 16, 65. [Google Scholar] [CrossRef]
- Mahyad, B.; Janfaza, S.; Hosseini, E.S. Bio-nano hybrid materials based on bacteriorhodopsin: Potential applications and future strategies. Adv. Colloid Interface Sci. 2015, 225, 194–202. [Google Scholar] [CrossRef]
- Liu, Y.; Turner, A.P.F.; Zhao, M.; Mak, W.C. Processable enzyme-hybrid conductive polymer composites for electrochemical biosensing. Biosens. Bioelectron. 2018, 100, 374–381. [Google Scholar] [CrossRef]
- Eguílaz, M.; Villalonga, R.; Rivas, G. Electrochemical biointerfaces based on carbon nanotubes-mesoporous silica hybrid material: Bioelectrocatalysis of hemoglobin and biosensing applications. Biosens. Bioelectron. 2018, 111, 144–151. [Google Scholar] [CrossRef]
- Lv, Z.; Wang, Y.; Chen, Z.; Sun, L.; Wang, J.; Chen, M.; Xu, Z.; Liao, Q.; Zhou, L.; Chen, X.; et al. Phototunable Biomemory Based on Light-Mediated Charge Trap. Adv. Sci. 2018, 5, 1800714. [Google Scholar] [CrossRef] [Green Version]
- Kramer, B.P.; Fischer, C.; Fussenegger, M. BioLogic gates enable logical transcription control in mammalian cells. Biotechnol. Bioeng. 2004, 87, 478–484. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.-W.; Oh, B.-K.; Kim, Y.J. Protein-based biomemory device consisting of the cysteine-modified azurin. Appl. Phys. Lett. 2007, 91, 263902. [Google Scholar] [CrossRef]
- Güzel, R.; Ersöz, A.; Dolak, I.; Say, R. Multistate proteinous biomemory device based on redox controllable hapten cross-linker. Mater. Sci. Eng. C 2017, 79, 336–342. [Google Scholar] [CrossRef]
- Min, J.; Kim, S.-U.; Kim, Y.J.; Yea, C.-H.; Choi, J.-W. Fabrication of Recombinant Azurin Self-assembled Layer for the Application of Bioelectronic Device. J. Nanosci. Nanotechnol. 2008, 8, 4982–4987. [Google Scholar] [CrossRef]
- Lee, T.; Yagati, A.K.; Min, J.; Choi, J.-W. Bioprocessing Device Composed of Protein/DNA/Inorganic Material Hybrid. Adv. Funct. Mater. 2014, 24, 1781–1789. [Google Scholar] [CrossRef]
- Kang, P.; Wang, M.C.; Nam, S.W. Bioelectronics with two-dimensional materials. Microelectron. Eng. 2016, 161, 18–35. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Liu, J.; Wang, C.; Leng, X.; Xiao, Y.; Fu, L. Synthesis of graphene and related two-dimensional materials for bioelectronics devices. Biosens. Bioelectron. 2017, 89, 28–42. [Google Scholar] [CrossRef]
- Choi, C.; Lee, Y.; Cho, K.W.; Koo, J.H.; Kim, D.-H. Wearable and Implantable Soft Bioelectronics Using Two-Dimensional Materials. Acc. Chem. Res. 2019, 52, 73–81. [Google Scholar] [CrossRef]
- Dunn, K.E.; Trefzer, M.A.; Johnson, S.; Tyrrell, A.M. Towards a Bioelectronic Computer: A Theoretical Study of a Multi-Layer Biomolecular Computing System That Can Process Electronic Inputs. Int. J. Mol. Sci. 2018, 19, 2620. [Google Scholar] [CrossRef]
- Katz, E. Biocomputing- tools, aims, perspectives. Curr. Opin. Biotechnol. 2015, 34, 202–208. [Google Scholar] [CrossRef]
- Choi, J.-W.; Kim, J.S.; Kim, S.-U.; Min, J. Charge Storage in Redox-active Azurin Monolayer on 11-MUA Modified Gold Surface. Biochip J. 2009, 3, 157–163. [Google Scholar]
- Güzel, R.; Ersöz, A.; Ziyadanoğulları, R.; Say, R. Nano-hemoglobin film based sextet state biomemory device by cross-linked photosensitive hapten monomer. Talanta 2018, 176, 85–91. [Google Scholar] [CrossRef]
- Hwang, H.J.; Carey, J.R.; Brower, E.T.; Gengenbach, A.J.; Abramite, J.A.; Lu, Y. Blue Ferrocenium Azurin: An Organometalloprotein with Tunable Redox Properties. J. Am. Chem. Soc. 2005, 127, 15356–15357. [Google Scholar] [CrossRef]
- Liu, J.; Chakraborty, S.; Hosseinzadeh, P.; Yu, Y.; Tian, S.; Petrik, I.; Bhagi, A.; Lu, Y. Metalloproteins Containing Cytochrome, Iron–Sulfur, or Copper Redox Centers. Chem. Rev. 2014, 114, 4366–4469. [Google Scholar] [CrossRef]
- Suárez, G.; Santschi, C.; Martin, O.J.F.; Slaveykova, V.I. Biosensor based on chemically-designed anchorable cytochrome c for the detection of H2O2 released by aquaticcells. Biosens. Bioelectron. 2013, 42, 385–390. [Google Scholar] [CrossRef]
- Artés, J.M.; Díez-Pérez, I.; Gorostiza, P. Transistor-like Behavior of Single Metalloprotein Junctions. Nano Lett. 2012, 12, 2679–2684. [Google Scholar] [CrossRef]
- Lee, T.; Kim, S.-U.; Min, J.; Choi, J.-W. Multilevel Biomemory Device Consisting of Recombinant Azurin/Cytochrome c. Adv. Mater. 2010, 22, 510–514. [Google Scholar] [CrossRef]
- Balabushevich, N.G.; Sholina, E.A.; Mikhalchik, E.V.; Filatova, L.Y.; Vikulina, A.S.; Volodkin, D. Self-Assembled Mucin-Containing Microcarriers via Hard Templating on CaCO3 Crystals. Micromachines 2018, 9, 307. [Google Scholar] [CrossRef]
- Norris, K.; Mishukova, O.I.; Zykwinska, A.; Colliec-Jouault, S.; Sinquin, C.; Koptioug, A.; Cuenot, S.; Kerns, J.G.; Surmeneva, M.A.; Surmenev, R.A.; Douglas, T.E.L. Marine Polysaccharide-Collagen Coatings on Ti6Al4V Alloy Formed by Self-Assembly. Micromachines 2019, 10, 68. [Google Scholar] [CrossRef]
- Jensen, P.S.; Chi, Q.; Zhang, J.; Ulstrup, J. Long-Range Interfacial Electrochemical Electron Transfer of Pseudomonas aeruginosa Azurin−Gold Nanoparticle Hybrid Systems. J. Phys. Chem. C 2009, 113, 13993–14000. [Google Scholar] [CrossRef]
- Cho, B.; Song, S.; Ji, Y.; Kim, T.-W.; Lee, T. Organic Resistive Memory Devices: Performance Enhancement, Integration, and Advanced Architectures. Adv. Funct. Mater. 2011, 21, 2806–2829. [Google Scholar] [CrossRef]
- Li, L.; Li, G. High-Performance Resistance-Switchable Multilayers of Graphene Oxide Blended with 1,3,4-Oxadiazole Acceptor Nanocomposite. Micromachines 2019, 10, 140. [Google Scholar] [CrossRef]
- Li, L. Tunable Memristic Characteristics Based on Graphene Oxide Charge-Trap Memory. Micromachines 2019, 10, 151. [Google Scholar] [CrossRef]
- Lee, T.; Yagati, A.K.; Pi, F.; Sharma, A.; Choi, J.-W.; Guo, P. Construction of RNA-Quantum Dot Chimera for Nanoscale Resistive Biomemory Application. ACS Nano 2015, 9, 6675–6682. [Google Scholar] [CrossRef]
- Shu, D.; Shu, Y.; Haque, F.; Abdelmawla, S.; Guo, P. Thermodynamically Stable RNA three-way junctions as platform for constructing multi-functional nanoparticles for delivery of therapeutics. Nat. Nanotechnol. 2011, 6, 658–667. [Google Scholar] [CrossRef]
- Yoon, J.; Mohammadniaei, M.; Choi, H.K.; Shin, M.; Bharate, B.G.; Lee, T.; Choi, J.-W. Resistive switching biodevice composed of MoS2-DNA heterolayer on the gold electrode. Appl. Surf. Sci. 2019, 478, 134–141. [Google Scholar] [CrossRef]
- Lembke, D.; Bertolazzi, S.; Kis, A. Single-Layer MoS2 Electronics. Acc. Chem. Res. 2015, 48, 100–110. [Google Scholar] [CrossRef]
- Barua, S.; Dutta, H.S.; Gogoi, S.; Devi, R.; Khan, R. Nanostructured MoS2-Based Advanced Biosensors: A Review. ACS Appl. Nano Mater. 2018, 1, 2–25. [Google Scholar] [CrossRef]
- Andersson, M.; Sinks, L.E.; Hayes, R.T.; Zhao, Y.; Wasielewski, M.R. Bio-Inspired Optically Controlled Ultrafast Molecular AND Gate. Angew. Chem. Int. Ed. 2003, 42, 3139–3143. [Google Scholar] [CrossRef]
- Sivan, S.; Tuchman, S.; Lotan, N. A biochemical logic gate using an enzyme and its inhibitor. Part II: The logic gate. BioSystems 2003, 70, 21–33. [Google Scholar] [CrossRef]
- Dung, N.Q.; Patil, D.; Duong, T.-T.; Jung, H.; Kim, D.; Yoon, S.-G. An amperometric glucose biosensor based on a GOx-entrapped TiO2–SWCNT composite. Sens. Actuator B-Chem. 2012, 166–167, 103–109. [Google Scholar] [CrossRef]
- Chen, Q.; Yoo, S.-Y.; Chung, Y.-H.; Lee, J.-Y.; Min, J.; Choi, J.-W. Control of electrochemical signals from quantum dots conjugated to organic materials by using DNA structure in an analog logic gate. Bioelectrochemistry 2016, 111, 1–6. [Google Scholar] [CrossRef]
- Chung, Y.-H.; Lee, T.; Yoo, S.-Y.; Min, J.; Choi, J.-W. Electrochemical Bioelectronic Device Consisting of Metalloprotein for Analog Decision Making. Sci. Rep. 2015, 5, 14501. [Google Scholar] [CrossRef] [Green Version]
- Gurunathan, S.; Klinman, D.M.; Seder, R.A. DNA Vaccines: Immunology, Application, and Optimization. Annu. Rev. Immunol. 2000, 18, 927–974. [Google Scholar] [CrossRef] [Green Version]
- França, L.T.; Carrilho, E.; Kist, T.B. A review of DNA sequencing techniques. Q. Rev. Biophys. 2002, 35, 169–200. [Google Scholar] [CrossRef]
- Pei, H.; Lu, N.; Wen, Y.; Song, S.; Liu, Y.; Yan, H.; Fan, C. A DNA Nanostructure-based Biomolecular Probe Carrier Platform for Electrochemical Biosensing. Adv. Mater. 2010, 22, 4754–4758. [Google Scholar] [CrossRef] [Green Version]
- Ono, A.; Cao, S.; Togashi, H.; Tashiro, M.; Fujimoto, T.; Machinami, T.; Oda, S.; Miyake, Y.; Okamotoa, I.; Tanaka, Y. Specific interactions between silver(I) ions and cytosine–cytosine pairs in DNA duplexes. Chem. Commun. 2008, 39, 4825–4827. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Zhang, L.; Liang, R.P.; Qiu, J.D. DNA Electronic: Logic Gates Based on Metal-Ion-Dependent Induction of Oligonucleotide Structural Motifs. Chem. Eur. J. 2013, 19, 6961–6965. [Google Scholar] [CrossRef]
- Guz, N.; Fedotova, T.A.; Fratto, B.E.; Schlesinger, O.; Alfonta, L.; Kolpashchikov, D.M.; Katz, E. Bioelectronic Interface Connecting Reversible Logic Gates Based on Enzyme and DNA Reactions. ChemPhysChem 2016, 17, 2247–2255. [Google Scholar] [CrossRef]
- Katz, E.; Minko, S. Enzyme-based logic systems interfaced with signal-responsive materials and electrodes. Chem. Commun. 2015, 51, 3493–3500. [Google Scholar] [CrossRef]
- Katz, E.; Poghossian, A.; Schöning, M.J. Enzyme-based logic gates and circuits-analytical applications and interfacing with electronics. Anal. Bioanal. Chem. 2017, 409, 81–94. [Google Scholar] [CrossRef]
- Muramatsu, S.; Kinbara, K.; Taguchi, H.; Ishii, N.; Aida, T. Semibiological Molecular Machine with an Implemented “AND” Logic Gate for Regulation of Protein Folding. J. Am. Chem. Soc. 2006, 128, 3764–3769. [Google Scholar] [CrossRef]
- Poghossian, A.; Katz, E.; Schöning, M.J. Enzyme logic AND-Reset and OR-Reset gates based on a field-effect electronic transducer modified with multi-enzyme membrane. Chem. Commun. 2015, 51, 6564–6567. [Google Scholar] [CrossRef]
- Fredkin, E.; Toffoli, T. Conservative Logic. In Collision-Based Computing; Adamatzky, A., Ed.; Springer: London, UK, 2002; pp. 47–81. [Google Scholar]
- O’Brien, J.L.; Pryde, G.J.; White, A.G.; Ralph, T.C.; Branning, D. Demonstration of an all-optical quantum controlled-NOT gate. Nature 2003, 426, 264–267. [Google Scholar] [CrossRef] [Green Version]
- Sun, D.-M.; Timmermans, M.Y.; Kaskela, A.; Nasibulin, A.G.; Kishimoto, S.; Mizutani, T.; Kauppinen, E.I.; Ohno, Y. Mouldable all-carbon integrated circuits. Nat. Commun. 2013, 4, 2302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, T.W.; Baude, P.F.; Gerlach, C.; Ender, D.E.; Muyres, D.; Haase, M.A.; Vogel, D.E.; Theiss, S.D. Recent Progress in Organic Electronics: Materials, Devices, and Processes. Chem. Mater. 2004, 16, 4413–4422. [Google Scholar] [CrossRef]
- Zou, C.; Wei, C.; Zhang, Q.; Liu, C.; Zhou, C.; Liu, Y. Four-Analog Computation Based on DNA Strand Displacement. ACS Omega 2017, 2, 4143–4160. [Google Scholar] [CrossRef]
- Son, M.; Park, T.H. The bioelectronic nose and tongue using olfactory and taste receptors: Analytical tools for food quality and safety assessment. Biotechnol. Adv. 2018, 36, 371–379. [Google Scholar] [CrossRef]
- Lim, J.-H.; Park, J.; Ahn, J.H.; Jin, H.J.; Hong, S.; Park, T.H. A peptide receptor-based bioelectronic nose for the real-time determination of seafood quality. Biosens. Bioelectron. 2013, 39, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Szaciłowski, K. Digital Information Processing in Molecular Systems. Chem. Rev. 2008, 108, 3481–3548. [Google Scholar] [CrossRef] [PubMed]
- Toriello, N.M.; Douglas, E.S.; Thaitrong, N.; Hsiao, S.C.; Francis, M.B.; Bertozzi, C.R.; Mathies, R.A. Integrated microfluidic bioprocessor for single-cell gene expression analysis. Proc. Natl. Acad. Sci. USA 2008, 105, 20173–20178. [Google Scholar] [CrossRef] [Green Version]
- Evans, A.C.; Thadani, N.N.; Suh, J. Biocomputing nanoplatforms as therapeutics and diagnostics. J. Controll. Release 2016, 240, 387–393. [Google Scholar] [CrossRef] [Green Version]
- Yurke, B.; Turberfield, A.J.; Mills, A.P., Jr.; Simmel, F.C.; Neumann, J.L. A DNA-fuelled molecular machine made of DNA. Nature 2000, 406, 605–608. [Google Scholar] [CrossRef]
- MacConnell, A.B.; Price, A.K.; Paegel, B.M. An Integrated Microfluidic Processor for DNA-Encoded Combinatorial Library Functional Screening. ACS Comb. Sci. 2017, 19, 181–192. [Google Scholar] [CrossRef]
- Shu, J.-J.; Wang, Q.-W.; Yong, K.-Y.; Shao, F.; Lee, K.J. Programmable DNA-Mediated Multitasking Processor. J. Phys. Chem. B 2015, 119, 5639–5644. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Jeerapan, I.; Sempionatto, J.R.; Barfidokht, A.; Mishra, R.K.; Campbell, A.S.; Hubble, L.J.; Wang, J. Wearable Bioelectronics: Enzyme-Based Body-Worn Electronic Devices. Acc. Chem. Res. 2018, 51, 2820–2828. [Google Scholar] [CrossRef]
- Lopes, P.A.; Paisana, H.; De Almeida, A.T.; Majidi, C.; Tavakoli, M. Hydroprinted Electronics: Ultrathin Stretchable Ag-In-Ga E-Skin for Bioelectronics and Human-Machine Interaction. ACS Appl. Mater. Interfaces 2018, 10, 38760–38768. [Google Scholar] [CrossRef]
- Nawroth, J.C.; Lee, H.; Feinberg, A.W.; Ripplinger, C.M.; McCain, M.L.; Grosberg, A.; Dabiri, J.O.; Parker, K.K. A tissue-engineered jellyfish with biomimetic propulsion. Nat. Biotechnol. 2012, 30, 792–797. [Google Scholar] [CrossRef]
- Williams, B.W.; Anand, S.V.; Rajagopalan, J.; Saif, M.; Taher, A. A self-propelled biohybrid swimmer at low Reynolds number. Nat. Commun. 2014, 5, 3081. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.R.; Zihlmann, C.; Akbari, M.; Assawes, P.; Cheung, L.; Zhang, K.; Manoharan, V.; Zhang, Y.S.; Yüksekkaya, M.; Wan, K.-T.; Nikkhah, M.; Dokmeci, M.R.; Tang, X.S.; Khademhosseini, A. Reduced Graphene Oxide-GelMA Hybrid Hydrogels as Scaffolds for Cardiac Tissue Engineering. Small 2016, 12, 27. [Google Scholar] [CrossRef]
- Olofsson, P.S.; Tracey, K.J. Bioelectronic medicine: Technology targeting molecular mechanisms for therapy. J. Intern. Med. 2017, 282, 3–4. [Google Scholar] [CrossRef]
- Löffler, S.; Melican, K.; Nilsson, K.P.R.; Richter-Dahlfors, A. Organic bioelectronics in medicine. J. Intern. Med. 2017, 282, 24–36. [Google Scholar] [CrossRef] [Green Version]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, J.; Lee, T.; Choi, J.-W. Development of Bioelectronic Devices Using Bionanohybrid Materials for Biocomputation System. Micromachines 2019, 10, 347. https://doi.org/10.3390/mi10050347
Yoon J, Lee T, Choi J-W. Development of Bioelectronic Devices Using Bionanohybrid Materials for Biocomputation System. Micromachines. 2019; 10(5):347. https://doi.org/10.3390/mi10050347
Chicago/Turabian StyleYoon, Jinho, Taek Lee, and Jeong-Woo Choi. 2019. "Development of Bioelectronic Devices Using Bionanohybrid Materials for Biocomputation System" Micromachines 10, no. 5: 347. https://doi.org/10.3390/mi10050347