Development of a Portable SPR Sensor for Nucleic Acid Detection
Abstract
:1. Introduction
2. Materials and Methods
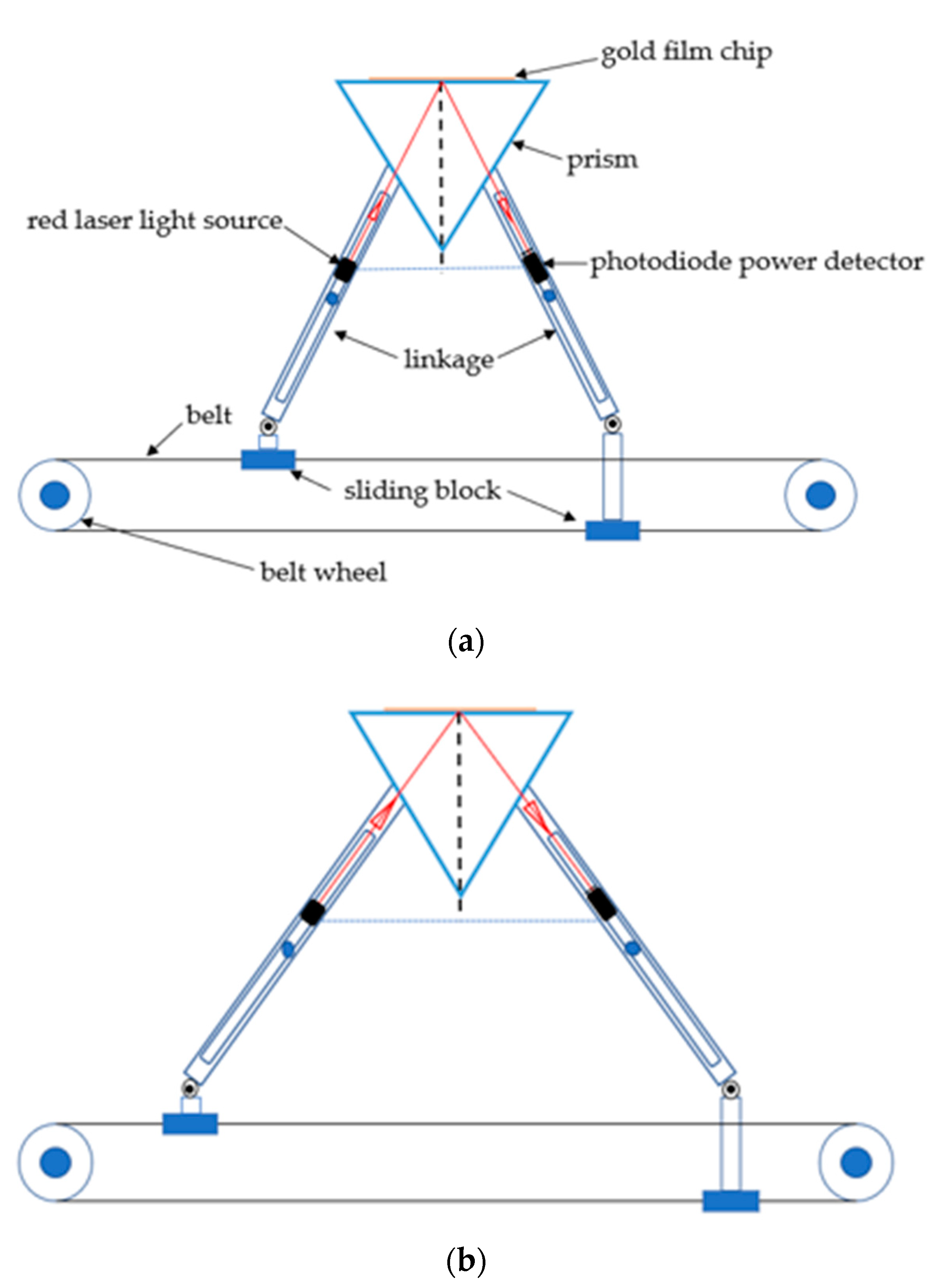
2.1. Surface Plasmon Resonance (SPR) Sensor Setup
2.2. SPR Chip and the Flow Cell
2.3. Materials and Samples
2.4. Sensor Performance Test
2.5. Experimental Protocol
3. Results and Discussion
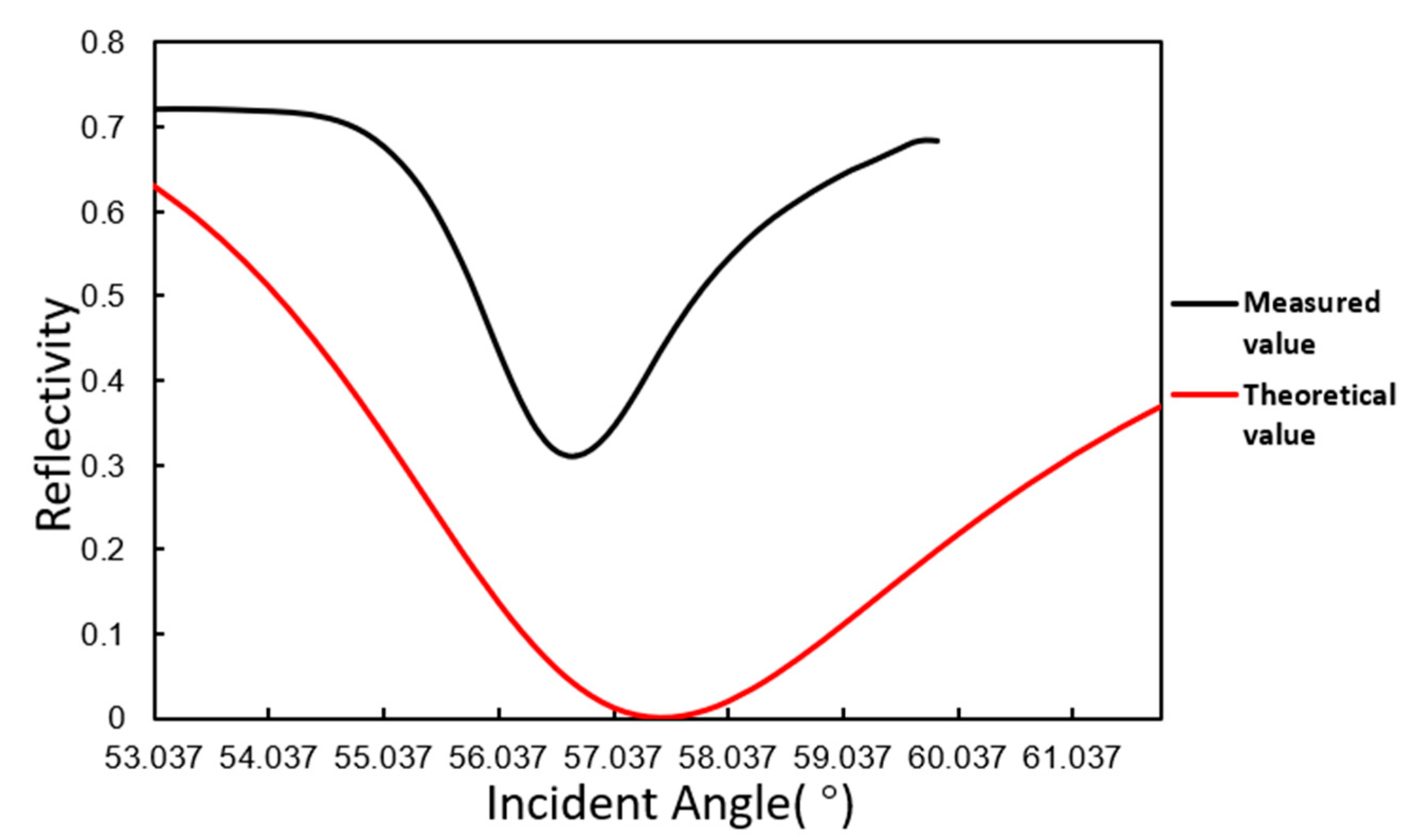
3.1. Angle Scanning Test
3.2. Refractive Index Resolution Test
3.2.1. Unmodified Gold Film Chip
3.2.2. Gold Film Chip Modified by BSA
3.3. DNA Probes Molecule Fixation
3.4. DNA Hybridization Detection
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Engvall, E.; Perlmann, P. Enzyme-linked immunosorbent assay (ELISA): Quantitative assay of immunoglobulin G. Immunochemistry 1971, 8, 871–874. [Google Scholar] [CrossRef]
- Balsari, A.; Poli, G.; Molina, V. ELISA for toxoplasma antibody detection:a comparison with other serodiagnostic tests. J. Clin. Pathol. 1980, 337, 640–643. [Google Scholar] [CrossRef] [Green Version]
- Huertas Cesar, S.; Calvo-Lozano, O.; Mitchell, A.; Lechuga, L. Advanced Evanescent-Wave Optical Biosensors for the Detection of Nucleic Acids: An Analytic Perspective. Front. Chem. 2019, 7, 2–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Kannegulla, A.; Wu, B.; Cheng, L.J. Quantum Dot-Fullerene Based Molecular Beacon Nanosensors for Rapid, Highly Sensitive Nucleic Acid Detection. ACS Appl. Mater. Interfaces 2018, 10, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Simona, S.; Laura, E.M.; Michela, S.M.; Marco, M.; Patrizia, B.; Maria, M. Simultaneous detection of transgenic DNA by surface plasmon resonance imaging with potential application to gene doping detection. Anal. Chem. 2011, 16, 6245–6253. [Google Scholar]
- Shoaie, N.; Forouzandeh, M.; Omidfar, K. Highly Sensitive Electrochemical Biosensor Based on Polyaniline and Gold Nanoparticles for DNA Detection. IEEE Sens. J. 2018, 18, 1835–1843. [Google Scholar] [CrossRef]
- Kostrzynska, M.; Bachand, A. Application of DNA microarray technology for detection, identification, and characterization of food-borne pathogens. Can. J. Microbiol. 2006, 52, 1–8. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Petz, L.N.; Melanson, V.R.; McMenamy, S.S.; Turell, J.M.; Long, L.S.; Pisarcik, S.E.; Kengluecha, A.; Jaichapor, B.; O’Guinn, M.L.; et al. Evaluation of a field-portable DNA microarray platform and nucleic Acid amplification strategies for the detection of arboviruses, arthropods, and bloodmeals. Am. J. Trop. Med. Hyg. 2013, 88, 245–253. [Google Scholar] [CrossRef] [Green Version]
- Maffert, P.; Reverchon, S.; Nasser, W.; Rozand, C.; Abaibou, H. New nucleic acid testing devices to diagnose infectious diseases in resource-limited settings. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1717–1731. [Google Scholar] [CrossRef]
- Wang, L.Y.; Chen, P.R.; Zheng, G.W.; Qi, N.; Yang, K. Research progress on detection methods of SARS-CoV-2. Drugs Clinic 2020, 35, 411–415. [Google Scholar]
- Gu, Y.; Tan, Y.J.; Wang, C.Y.; Nie, J.L.; Yu, J.R.; Li, Y.H. A Surface Plasmon Resonance Sensor Platform Coupled with Gold Nanoparticle Probes for Unpurified Nucleic Acids Detection. Anal. Lett. 2012, 45, 2210–2220. [Google Scholar] [CrossRef]
- Mahmoudpour, M.; Dolatabadi, J.E.N.; Torbati, M.; Homayouni-Rad, A. Nanomaterials based surface plasmon resonance signal enhancement for detection of environmental pollutions. Biosens. Bioelectron. 2019, 127, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Gorodkiewicz, E.; Lukaszewski, Z. Recent Progress in Surface Plasmon Resonance Biosensors (2016 to Mid-2018). Biosensors 2018, 8, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Homola, J. Surface Plasmon Resonance Sensors for Detection of Chemical and Biological Species. Chem. Rev. 2008, 108, 462–493. [Google Scholar] [CrossRef]
- Sin, L.Y.M.; Mach, E.K.; Wong, P.K.; Liao, J.C. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 2014, 14, 225–244. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.J.; Chen, H.W.; Song, D.Q.; Mou, Y.; Zhang, H.Q.; Jin, Q.H. Surface plasmon resonance sensor, part I: Fundamentals. Expert Rev. Mol. Diagn. 2000, 25, 79–80. [Google Scholar]
- Chen, Y.; Cui, D.F.; Cai, H.Y.; Wang, J.B.; Wang, Y.J. Design of mini SPR biochemical sensor. Transducer Mierosystem Technol. 2006, 25, 79–80. [Google Scholar]
- Soelberg, S.D.; Chinowsky, T.; Geiss, G.; Spinelli, C.B.; Stevens, R.; Steve, N.; Kauffman, P.; Yee, S.; Furlong, C.E. A portable surface plasmon resonance sensor system for real-time monitoring of small to large analytes. Environ. Technol. 2005, 32, 669–670. [Google Scholar] [CrossRef]
- Tang, X.Y.; Zhang, H.Y. Advances in the Application of SPR Biosens. China Anim. Husb. Vet. Med. 2006, 33, 47–50. [Google Scholar]
- Maisonneuve, M.; Valsecchi, C.; Wang, C.G.; Brolo, A.; Michel, M. Leukemic marker detection using a spectro-polarimetric surface plasmon resonance platform. Biosens. Bioelectron. 2015, 63, 80–85. [Google Scholar] [CrossRef]
- Wang, Q.; Jing, J.Y.; Wang, B.T. Highly Sensitive SPR Biosensor Based on Graphene Oxide and Staphylococcal Protein A Co-Modified TFBG for Human IgG Detection. IEEE Trans. Instrum. Meas. 2019, 68, 3350–3357. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.M.; Wang, J.; Fu, W.L.; Yao, C.Y. A SPR biosensor based on signal amplification using antibody-QD conjugates for quantitative determination of multiple tumor markers. Sci. Rep.-UK. 2016, 6, 33140. [Google Scholar] [CrossRef]
- Liu, Y. Investigation of Miniaturized Optical Fiber Surface Plasmon Resonance Biological Sensor Systems. Ph.D. Thesis, Dalian University of Technology, Dalian, China, 2016. [Google Scholar]
- Huang, Y. Research on Refractive Index Measurement System Based on SPR Using Wavelength Modulation. Ph.D. Thesis, Heilongjiang University, Heilongjiang, China, 2012. [Google Scholar]
- Liu, X.C. Study on Angle Modulated Surface Plasmon Resonance Spectrometer. Master’s Thesis, Dalian University of Technology, Dalian, China, 2015. [Google Scholar]
- Qi, P.; Li, Y.; Li, S.P.; Zhong, J.G. Portable Surface Plasmon Resonance Biosensor and Its Applications. In Proceedings of the 14th International Conference on Photonics and Imaging in Biology and Medicine, Suzhou, China, 26–28 September 2017. [Google Scholar]
- Laksono, F.D.; Supardianningsih; Arifin, M.; Abraha, K. Development of low cost and accurate homemade sensor system based on Surface Plasmon Resonance (SPR). In Proceedings of the 5th International Conference on Theoretical and Applied Physics, Rome, Italy, 6–8 September 2018. [Google Scholar]
- Somarapalli, M.; Jolivot, R.; Mohammed, W. Realization of Low-Cost Multichannel Surface Plasmon Resonance Based Optical Transducer. Photonic Sens. 2018, 8, 289–302. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.G.; Chen, J.R.; Liu, R.C.; Wang, Y.L. Low Cost Surface Plasmon Resonance Sensor System. Instrum. Tech. Sens. 2019, 5, 51–53. [Google Scholar]
- Vestri, A.; Margheri, G.; Landini, E.; Meacci, E.; Tiribilli, B. A versatile and compact surface plasmon resonance spectrometer based on single board computer. Rev. Sci. Instrum. 2020, 91, 0131061–0131067. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.A.; Kim, S.J.; Byun, K.M.; Lee, S.H.; Park, T.H.; Kim, S.G.; Shuler, M.L. Avian Influenza-DNA Hybridization Detection using Wavelength Interrogation-based Surface Plasmon Resonance Biosensor. In Proceedings of the 8th Annual IEEE Conference on Sensors, Christchurch, New Zealand, 25–28 October 2009. [Google Scholar]
- Ding, X.J.; Yan, Y.R.; Li, S.Q.; Zhang, Y.; Cheng, W.; Cheng, Q.; Ding, S.J. Surface plasmon resonance biosensor for highly sensitive detection of microRNA based on DNA super-sandwich assemblies and streptavidin signal amplification. Anal. Chim. Acta 2015, 874, 60–64. [Google Scholar] [CrossRef]
- Bagi, A.; Soelberg, S.D.; Furlong, C.E.; Baussant, T. Implementing Morpholino-Based Nucleic Acid Sensing on a Portable Surface Plasmon Resonance Instrument for Future Application in Environmental Monitoring. Sensors 2018, 18, 3259. [Google Scholar] [CrossRef] [Green Version]
- Sípová, H.; Homola, J. Surface plasmon resonance sensing of nucleic acids: A review. Anal. Chim. Acta 2013, 773, 9–23. [Google Scholar] [CrossRef]
- Liu, X.Y.; Bai, Y.Q.; Wang, C.Y.; Dai, Z.Q.; Li, Y.H. Oligonucleotide Hybridization Detection Based on Surface Plasmon Resonance Technology. Space Med. Med. Eng. 2007, 20, 18. [Google Scholar]







© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Zhang, L.; Zhang, H.; Li, Y.; Liu, L.; Chen, Y.; Qiu, X.; Yu, D. Development of a Portable SPR Sensor for Nucleic Acid Detection. Micromachines 2020, 11, 526. https://doi.org/10.3390/mi11050526
Huang Y, Zhang L, Zhang H, Li Y, Liu L, Chen Y, Qiu X, Yu D. Development of a Portable SPR Sensor for Nucleic Acid Detection. Micromachines. 2020; 11(5):526. https://doi.org/10.3390/mi11050526
Chicago/Turabian StyleHuang, Yafeng, Lulu Zhang, Hao Zhang, Yichen Li, Luyao Liu, Yuanyuan Chen, Xianbo Qiu, and Duli Yu. 2020. "Development of a Portable SPR Sensor for Nucleic Acid Detection" Micromachines 11, no. 5: 526. https://doi.org/10.3390/mi11050526



