Developments in FRET- and BRET-Based Biosensors
Abstract
:1. Introduction
1.1. FRET and FRET Systems
1.2. BRET and BRET Systems
2. FRET and BRET Strategies in Biosensors
2.1. Biosensors for Biomedical Research
2.1.1. Biosensors for Bioassay and Diagnosis
2.1.2. Biosensors for In Vivo Imaging
2.2. Biosensors for Environmental Applications
2.3. Biosensors for In Vivo Dynamic Analysis of Metabolic Flux
3. Conclusions and Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Förster, T. Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann. Der Phys. 1948, 437, 55–75. [Google Scholar] [CrossRef]
- Wu, P.; Brand, L. Resonance energy transfer: Methods and applications. Anal. Biochem. 1994, 218, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pfleger, K.D.; Seeber, R.M.; Eidne, K.A. Bioluminescence resonance energy transfer (BRET) for the real-time detection of protein-protein interactions. Nat. Protoc. 2006, 1, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Algar, W.R.; Hildebrandt, N.; Vogel, S.S.; Medintz, I.L. FRET as a biomolecular research tool—Understanding its potential while avoiding pitfalls. Nat. Methods 2019, 16, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Salahpour, A.; Espinoza, S.; Masri, B.; Lam, V.; Barak, L.S.; Gainetdinov, R.R. BRET biosensors to study GPCR biology, pharmacology, and signal transduction. Front. Endocrinol. 2012, 3, 105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jares-Erijman, E.A.; Jovin, T.M. FRET imaging. Nat. Biotechnol. 2003, 21, 1387–1395. [Google Scholar] [CrossRef]
- Imani, M.; Mohajeri, N.; Rastegar, M.; Zarghami, N. Recent advances in FRET-Based biosensors for biomedical applications. Anal. Biochem. 2021, 630, 114323. [Google Scholar] [CrossRef]
- Stryer, L.; Haugland, R.P. Energy transfer: A spectroscopic ruler. Proc. Natl. Acad. Sci. USA 1967, 58, 719–726. [Google Scholar] [CrossRef] [Green Version]
- Deal, J.; Pleshinger, D.J.; Johnson, S.C.; Leavesley, S.J.; Rich, T.C. Milestones in the development and implementation of FRET-based sensors of intracellular signals: A biological perspective of the history of FRET. Cell Signal. 2020, 75, 109769. [Google Scholar] [CrossRef]
- Stryer, L. Fluorescence energy transfer as a spectroscopic ruler. Annu. Rev. Biochem. 1978, 47, 819–846. [Google Scholar] [CrossRef]
- Sapsford, K.E.; Berti, L.; Medintz, I.L. Materials for fluorescence resonance energy transfer analysis: Beyond traditional donor-acceptor combinations. Angew. Chem. Int. Ed. Engl. 2006, 45, 4562–4589. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Hu, M.; Zhan, P.; Peng, X. Energy transfer cassettes based on organic fluorophores: Construction and applications in ratiometric sensing. Chem. Soc. Rev. 2013, 42, 29–43. [Google Scholar] [CrossRef]
- Mattsson, L.; Wegner, K.D.; Hildebrandt, N.; Soukka, T. Upconverting nanoparticle to quantum dot FRET for homogeneous double-nano biosensors. RSC Adv. 2015, 5, 13270–13277. [Google Scholar] [CrossRef]
- Doughan, S.; Uddayasankar, U.; Krull, U.J. A paper-based resonance energy transfer nucleic acid hybridization assay using upconversion nanoparticles as donors and quantum dots as acceptors. Anal. Chim. Acta 2015, 878, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Prasher, D.C.; Eckenrode, V.K.; Ward, W.W.; Prendergast, F.G.; Cormier, M.J. Primary structure of the Aequorea victoria green-fluorescent protein. Gene 1992, 111, 229–233. [Google Scholar] [CrossRef]
- Chalfie, M.; Tu, Y.; Euskirchen, G.; Ward, W.W.; Prasher, D.C. Green fluorescent protein as a marker for gene expression. Science 1994, 263, 802–805. [Google Scholar] [CrossRef] [Green Version]
- Inouye, S.; Tsuji, F.I. Aequorea green fluorescent protein. FEBS Lett. 1994, 341, 277–280. [Google Scholar] [CrossRef] [Green Version]
- Heim, R.; Prasher, D.C.; Tsien, R.Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA 1994, 91, 12501–12504. [Google Scholar] [CrossRef] [Green Version]
- Miyawaki, A.; Llopis, J.; Heim, R.; McCaffery, J.M.; Adams, J.A.; Ikura, M.; Tsien, R.Y. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 1997, 388, 882–887. [Google Scholar] [CrossRef]
- Ormo, M.; Cubitt, A.B.; Kallio, K.; Gross, L.A.; Tsien, R.Y.; Remington, S.J. Crystal structure of the Aequorea victoria green fluorescent protein. Science 1996, 273, 1392–1395. [Google Scholar] [CrossRef]
- Cormack, B.P.; Valdivia, R.H.; Falkow, S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 1996, 173, 33–38. [Google Scholar] [CrossRef]
- Zolotukhin, S.; Potter, M.; Hauswirth, W.W.; Guy, J.; Muzyczka, N. A humanized green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J. Virol. 1996, 70, 4646–4654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heim, R.; Cubitt, A.B.; Tsien, R.Y. Improved green fluorescence. Nature 1995, 373, 663–664. [Google Scholar] [CrossRef] [PubMed]
- Griesbeck, O.; Baird, G.S.; Campbell, R.E.; Zacharias, D.A.; Tsien, R.Y. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 2001, 276, 29188–29194. [Google Scholar] [CrossRef] [Green Version]
- Nagai, T.; Ibata, K.; Park, E.S.; Kubota, M.; Mikoshiba, K.; Miyawaki, A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002, 20, 87–90. [Google Scholar] [CrossRef]
- Matz, M.V.; Fradkov, A.F.; Labas, Y.A.; Savitsky, A.P.; Zaraisky, A.G.; Markelov, M.L.; Lukyanov, S.A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999, 17, 969–973. [Google Scholar] [CrossRef]
- Baird, G.S.; Zacharias, D.A.; Tsien, R.Y. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA 2000, 97, 11984–11989. [Google Scholar] [CrossRef] [Green Version]
- Shaner, N.C.; Campbell, R.E.; Steinbach, P.A.; Giepmans, B.N.; Palmer, A.E.; Tsien, R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef]
- Mitra, R.D.; Silva, C.M.; Youvan, D.C. Fluorescence resonance energy transfer between blue-emitting and red-shifted excitation derivatives of the green fluorescent protein. Gene 1996, 173, 13–17. [Google Scholar] [CrossRef]
- Heim, R.; Tsien, R.Y. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 1996, 6, 178–182. [Google Scholar] [CrossRef]
- Nguyen, A.W.; Daugherty, P.S. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 2005, 23, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, N.; Spillmann, C.M.; Algar, W.R.; Pons, T.; Stewart, M.H.; Oh, E.; Susumu, K.; Diaz, S.A.; Delehanty, J.B.; Medintz, I.L. Energy Transfer with Semiconductor Quantum Dot Bioconjugates: A Versatile Platform for Biosensing, Energy Harvesting, and Other Developing Applications. Chem. Rev. 2017, 117, 536–711. [Google Scholar] [CrossRef] [PubMed]
- Shipunova, V.O.; Shilova, O.N.; Shramova, E.I.; Deyev, S.M.; Proshkina, G.M. A Highly Specific Substrate for NanoLUC Luciferase Furimazine Is Toxic in vitro and in vivo. Russ. J. Bioorganic Chem. 2018, 44, 225–228. [Google Scholar] [CrossRef]
- Orioka, M.; Eguchi, M.; Mizui, Y.; Ikeda, Y.; Sakama, A.; Li, Q.J.; Yoshimura, H.; Ozawa, T.; Citterio, D.; Hiruta, Y. A Series of Furimazine Derivatives for Sustained Live-Cell Bioluminescence Imaging and Application to the Monitoring of Myogenesis at the Single-Cell Level. Bioconjugate Chem. 2022, 33, 496–504. [Google Scholar] [CrossRef]
- Machleidt, T.; Woodroofe, C.C.; Schwinn, M.K.; Méndez, J.; Robers, M.B.; Zimmerman, K.; Otto, P.; Daniels, D.L.; Kirkland, T.A.; Wood, K.V. NanoBRET--A Novel BRET Platform for the Analysis of Protein-Protein Interactions. ACS Chem. Biol. 2015, 10, 1797–1804. [Google Scholar] [CrossRef]
- Sun, S.; Yang, X.; Wang, Y.; Shen, X. In Vivo Analysis of Protein-Protein Interactions with Bioluminescence Resonance Energy Transfer (BRET): Progress and Prospects. Int. J. Mol. Sci. 2016, 17, 1704. [Google Scholar] [CrossRef] [Green Version]
- Yeh, H.W.; Ai, H.W. Development and Applications of Bioluminescent and Chemiluminescent Reporters and Biosensors. Annu. Rev. Anal. Chem. 2019, 12, 129–150. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Piston, D.W.; Johnson, C.H. A bioluminescence resonance energy transfer (BRET) system: Application to interacting circadian clock proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 151–156. [Google Scholar] [CrossRef] [Green Version]
- Bertrand, L.; Parent, S.; Caron, M.; Legault, M.; Joly, E.; Angers, S.; Bouvier, M.; Brown, M.; Houle, B.; Ménard, L. The BRET2/arrestin assay in stable recombinant cells: A platform to screen for compounds that interact with G protein-coupled receptors (GPCRS). J. Recept. Signal Transduct. Res. 2002, 22, 533–541. [Google Scholar] [CrossRef]
- Kocan, M.; See, H.B.; Seeber, R.M.; Eidne, K.A.; Pfleger, K.D. Demonstration of improvements to the bioluminescence resonance energy transfer (BRET) technology for the monitoring of G protein-coupled receptors in live cells. J. Biomol. Screen 2008, 13, 888–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfleger, K.D.; Dromey, J.R.; Dalrymple, M.B.; Lim, E.M.; Thomas, W.G.; Eidne, K.A. Extended bioluminescence resonance energy transfer (eBRET) for monitoring prolonged protein-protein interactions in live cells. Cell Signal. 2006, 18, 1664–1670. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Soutto, M.; Xu, X.; Zhang, Y.; Johnson, C.H. Bioluminescence resonance energy transfer (BRET) imaging in plant seedlings and mammalian cells. Methods Mol. Biol. 2011, 680, 3–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshino, H.; Nakajima, Y.; Ohmiya, Y. Luciferase-YFP fusion tag with enhanced emission for single-cell luminescence imaging. Nat. Methods 2007, 4, 637–639. [Google Scholar] [CrossRef] [PubMed]
- De, A.; Ray, P.; Loening, A.M.; Gambhir, S.S. BRET3: A red-shifted bioluminescence resonance energy transfer (BRET)-based integrated platform for imaging protein-protein interactions from single live cells and living animals. FASEB J. 2009, 23, 2702–2709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dragulescu-Andrasi, A.; Chan, C.T.; De, A.; Massoud, T.F.; Gambhir, S.S. Bioluminescence resonance energy transfer (BRET) imaging of protein-protein interactions within deep tissues of living subjects. Proc. Natl. Acad. Sci. USA 2011, 108, 12060–12065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimri, S.; Basu, S.; De, A. Use of BRET to Study Protein-Protein Interactions In Vitro and In Vivo. Methods Mol. Biol. 2016, 1443, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Bae Kim, S.; Fujii, R.; Natarajan, A.; Massoud, T.F.; Paulmurugan, R. Ligand-activated BRET9 imaging for measuring protein-protein interactions in living mice. Chem. Commun. 2019, 56, 281–284. [Google Scholar] [CrossRef]
- Hiblot, J.; Yu, Q.; Sabbadini, M.D.B.; Reymond, L.; Xue, L.; Schena, A.; Sallin, O.; Hill, N.; Griss, R.; Johnsson, K. Luciferases with Tunable Emission Wavelengths. Angew. Chem. Int. Ed. Engl. 2017, 56, 14556–14560. [Google Scholar] [CrossRef]
- Kim, S.B.; Paulmurugan, R. Bioluminescent Imaging Systems for Assay Developments. Anal. Sci. 2021, 37, 233–247. [Google Scholar] [CrossRef]
- Constantinou, A.; Polizzi, K.M. Opportunities for bioprocess monitoring using FRET biosensors. Biochem. Soc. Trans. 2013, 41, 1146–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadoine, M.; Cerminara, M.; Gerrits, M.; Fitter, J.; Katranidis, A. Cotranslational Incorporation into Proteins of a Fluorophore Suitable for smFRET Studies. ACS Synth. Biol. 2018, 7, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hu, Y.; Yang, X.; Tang, Y.; Han, S.; Kang, A.; Deng, H.; Chi, Y.; Zhu, D.; Lu, Y. FÖrster resonance energy transfer (FRET)-based biosensors for biological applications. Biosens. Bioelectron. 2019, 138, 111314. [Google Scholar] [CrossRef] [PubMed]
- Chappe, Y.; Michel, P.; Joushomme, A.; Barbeau, S.; Pierredon, S.; Baron, L.; Garenne, A.; Poulletier De Gannes, F.; Hurtier, A.; Mayer, S.; et al. High-Throughput Screening of Transient Receptor Potential Channel 1 Ligands in the Light of the Bioluminescence Resonance Energy Transfer Technique. Mol. Pharmacol. 2021, 100, 237–257. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Liu, M.-h.; Zhang, C.-y. Integration of isothermal amplification with quantum dot- based fluorescence resonance energy transfer for simultaneous detection of multiple microRNAs. Chem. Sci. 2018, 9, 4258–4267. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Xing, X.; Zou, T.; Wang, Z.; Zhao, R.; Hong, P.; Peng, S.; Zhang, X.; Wang, Y. A novel and sensitive ratiometric fluorescence assay for carbendazim based on N-doped carbon quantum dots and gold nanocluster nanohybrid. J. Hazard. Mater. 2020, 386, 121958. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, F.; Zhang, H.; Zhang, Y.; Liu, M.; Liu, Y. Universal Ti3C2 MXenes Based Self-Standard Ratiometric Fluorescence Resonance Energy Transfer Platform for Highly Sensitive Detection of Exosomes. Anal. Chem. 2018, 90, 12737–12744. [Google Scholar] [CrossRef]
- Das, P.; Krull, U.J. Detection of a cancer biomarker protein on modified cellulose paper by fluorescence using aptamer-linked quantum dots. Analyst 2017, 142, 3132–3135. [Google Scholar] [CrossRef]
- Liu, X.; Hou, Y.; Chen, S.; Liu, J. Controlling dopamine binding by the new aptamer for a FRET-based biosensor. Biosens. Bioelectron. 2021, 173, 112798. [Google Scholar] [CrossRef]
- Singh, S.; Sharma, M.P.; Alqarawi, A.A.; Hashem, A.; Abd Allah, E.F.; Ahmad, A. Real-Time Optical Detection of Isoleucine in Living Cells through a Genetically-Encoded Nanosensor. Sensors 2020, 20, 146. [Google Scholar] [CrossRef]
- Chen, J.; Liu, J.; Wang, J.; Zhang, Y.; Wang, X.; Zhou, N. Fluorescent biosensor based on FRET and catalytic hairpin assembly for sensitive detection of polysialic acid by using a new screened DNA aptamer. Talanta 2022, 242, 123282. [Google Scholar] [CrossRef]
- Calamera, G.; Li, D.; Ulsund, A.H.; Kim, J.J.; Neely, O.C.; Moltzau, L.R.; Bjornerem, M.; Paterson, D.; Kim, C.; Levy, F.O.; et al. FRET-based cyclic GMP biosensors measure low cGMP concentrations in cardiomyocytes and neurons. Commun. Biol. 2019, 2, 394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Huo, F.; Cheng, F.; Yin, C. Employing an ICT-FRET Integration Platform for the Real-Time Tracking of SO2 Metabolism in Cancer Cells and Tumor Models. J. Am. Chem. Soc. 2020, 142, 6324–6331. [Google Scholar] [CrossRef] [PubMed]
- Crocker, H.; Pelosse, M.; Schlattner, U.; Berger, I. AMPfret: Synthetic nanosensor for cellular energy states. Biochem. Soc. Trans. 2020, 48, 103–111. [Google Scholar] [CrossRef]
- Weihs, F.; Anderson, A.; Trowell, S.; Caron, K. Resonance Energy Transfer-Based Biosensors for Point-of-Need Diagnosis-Progress and Perspectives. Sensors 2021, 21, 660. [Google Scholar] [CrossRef] [PubMed]
- Griss, R.; Schena, A.; Reymond, L.; Patiny, L.; Werner, D.; Tinberg, C.E.; Baker, D.; Johnsson, K. Bioluminescent sensor proteins for point-of-care therapeutic drug monitoring. Nat. Chem. Biol. 2014, 10, 598–603. [Google Scholar] [CrossRef]
- Arts, R.; den Hartog, I.; Zijlema, S.E.; Thijssen, V.; van der Beelen, S.H.; Merkx, M. Detection of Antibodies in Blood Plasma Using Bioluminescent Sensor Proteins and a Smartphone. Anal. Chem. 2016, 88, 4525–4532. [Google Scholar] [CrossRef]
- van Rosmalen, M.; Ni, Y.; Vervoort, D.F.M.; Arts, R.; Ludwig, S.K.J.; Merkx, M. Dual-Color Bioluminescent Sensor Proteins for Therapeutic Drug Monitoring of Antitumor Antibodies. Anal. Chem. 2018, 90, 3592–3599. [Google Scholar] [CrossRef]
- Arts, R.; Ludwig, S.K.J.; van Gerven, B.C.B.; Estirado, E.M.; Milroy, L.-G.; Merkx, M. Semisynthetic Bioluminescent Sensor Proteins for Direct Detection of Antibodies and Small Molecules in Solution. ACS Sens. 2017, 2, 1730–1736. [Google Scholar] [CrossRef]
- Tenda, K.; van Gerven, B.; Arts, R.; Hiruta, Y.; Merkx, M.; Citterio, D. Paper-Based Antibody Detection Devices Using Bioluminescent BRET-Switching Sensor Proteins. Angew. Chem. Int. Ed. Engl. 2018, 57, 15369–15373. [Google Scholar] [CrossRef]
- Ni, Y.; Arts, R.; Merkx, M. Ratiometric Bioluminescent Sensor Proteins Based on Intramolecular Split Luciferase Complementation. ACS Sens. 2019, 4, 20–25. [Google Scholar] [CrossRef]
- Takahashi, R.; Yasuda, T.; Ohmuro-Matsuyama, Y.; Ueda, H. BRET Q-Body: A Ratiometric Quench-based Bioluminescent Immunosensor Made of Luciferase-Dye-Antibody Fusion with Enhanced Response. Anal. Chem. 2021, 93, 7571–7578. [Google Scholar] [CrossRef]
- Yu, Q.; Xue, L.; Hiblot, J.; Griss, R.; Fabritz, S.; Roux, C.; Binz, P.A.; Haas, D.; Okun, J.G.; Johnsson, K. Semisynthetic sensor proteins enable metabolic assays at the point of care. Science 2018, 361, 1122–1126. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, L.; Ni, W.; Luo, Q.; Zhu, C.; Wu, Y. Portable and Field-Ready Detection of Circulating MicroRNAs with Paper-Based Bioluminescent Sensing and Isothermal Amplification. Anal. Chem. 2019, 91, 14838–14841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Cumberbatch, D.; Centanni, S.; Shi, S.Q.; Winder, D.; Webb, D.; Johnson, C.H. Coupling optogenetic stimulation with NanoLuc-based luminescence (BRET) Ca(++) sensing. Nat. Commun. 2016, 7, 13268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, S.R.; Harootunian, A.T.; Buechler, Y.J.; Taylor, S.S.; Tsien, R.Y. Fluorescence ratio imaging of cyclic AMP in single cells. Nature 1991, 349, 694–697. [Google Scholar] [CrossRef]
- Nikolaev, V.O.; Lohse, M.J. Monitoring of cAMP synthesis and degradation in living cells. Physiology 2006, 21, 86–92. [Google Scholar] [CrossRef]
- Zaccolo, M.; De Giorgi, F.; Cho, C.Y.; Feng, L.; Knapp, T.; Negulescu, P.A.; Taylor, S.S.; Tsien, R.Y.; Pozzan, T. A genetically encoded, fluorescent indicator for cyclic AMP in living cells. Nat. Cell Biol. 2000, 2, 25–29. [Google Scholar] [CrossRef]
- Zaccolo, M.; Pozzan, T. Discrete microdomains with high concentration of cAMP in stimulated rat neonatal cardiac myocytes. Science 2002, 295, 1711–1715. [Google Scholar] [CrossRef]
- Goaillard, J.M.; Vincent, P. Serotonin suppresses the slow afterhyperpolarization in rat intralaminar and midline thalamic neurones by activating 5-HT7 receptors. J. Physiol. Lond. 2002, 541, 453–465. [Google Scholar] [CrossRef]
- Webb, R.J.; Marshall, F.; Swann, K.; Carroll, J. Follicle-stimulating hormone induces a gap junction-dependent dynamic change in [cAMP] and protein kinase a in mammalian oocytes. Dev. Biol. 2002, 246, 441–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, N.; Kyozuka, K.; Deguchi, R. Increase in intracellular cAMP is a prerequisite signal for initiation of physiological oocyte meiotic maturation in the hydrozoan Cytaeis uchidae. Dev. Biol. 2006, 298, 248–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagai, Y.; Miyazaki, M.; Aoki, R.; Zama, T.; Inouye, S.; Hirose, K.; Iino, M.; Hagiwara, M. A fluorescent indicator for visualizing cAMP-induced phosphorylation in vivo. Nat. Biotechnol. 2000, 18, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Min, S.H.; French, A.R.; Trull, K.J.; Tat, K.; Varney, S.A.; Tantama, M. Ratiometric BRET Measurements of ATP with a Genetically-Encoded Luminescent Sensor. Sensors 2019, 19, 3502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shcherbakova, D.M.; Cammer, N.C.; Huisman, T.M.; Verkhusha, V.V.; Hodgson, L. Direct multiplex imaging and optogenetics of Rho GTPases enabled by near-infrared FRET. Nat. Chem. Biol. 2018, 14, 591–600. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Wang, J.; Lei, Z.; Lu, L.; Wang, S.; Zhang, H.; Li, B.; Zhang, F. NIR-II pH Sensor with a FRET Adjustable Transition Point for In Situ Dynamic Tumor Microenvironment Visualization. Angew. Chem. Int. Ed. 2021, 60, 5091–5095. [Google Scholar] [CrossRef]
- Taylor, A.; Sharkey, J.; Plagge, A.; Wilm, B.; Murray, P. Multicolour In Vivo Bioluminescence Imaging Using a NanoLuc-Based BRET Reporter in Combination with Firefly Luciferase. Contrast Media Mol. Imaging 2018, 2018, 2514796. [Google Scholar] [CrossRef] [Green Version]
- Tang, Z.; Liu, X.; Su, B.; Chen, Q.; Cao, H.; Yun, Y.; Xu, Y.; Hammock, B.D. Ultrasensitive and rapid detection of ochratoxin A in agro-products by a nanobody-mediated FRET-based immunosensor. J. Hazard. Mater. 2020, 387, 121678. [Google Scholar] [CrossRef]
- Sabet, F.S.; Hosseini, M.; Khabbaz, H.; Dadmehr, M.; Ganjali, M.R. FRET-based aptamer biosensor for selective and sensitive detection of aflatoxin B1 in peanut and rice. Food Chem. 2017, 220, 527–532. [Google Scholar] [CrossRef]
- Wu, X.L.; Song, Y.; Yan, X.; Zhu, C.Z.; Ma, Y.Q.; Du, D.; Lin, Y.H. Carbon quantum dots as fluorescence resonance energy transfer sensors for organophosphate pesticides determination. Biosens. Bioelectron. 2017, 94, 292–297. [Google Scholar] [CrossRef]
- Li, Y.; Ouyang, Q.; Li, H.H.; Chen, M.; Zhan, Z.Z.; Chen, Q.S. Turn-On Fluoresence Sensor for Hg2+ in Food Based on FRET between Aptamers-Functionalized Upconversion Nanoparticles and Gold Nanoparticles. J. Agric. Food Chem. 2018, 66, 6188–6195. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Feng, D.-Q.; Qian, Y.; Wang, W.; Zhu, J.-J. Construction of FRET biosensor for off-on detection of lead ions based on carbon dots and gold nanorods. Talanta 2019, 201, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Wang, S.; Lin, M.; Jin, Y.; Zhang, S.; Cui, X.; Gong, Y.; Li, A.; Xu, F.; Lu, T.J. Upconversion nanoparticles based FRET aptasensor for rapid and ultrasenstive bacteria detection. Biosens. Bioelectron. 2017, 90, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.N.; Ishida, R.; Hattori, M.; Matsuda, T.; Nagai, T. Bioluminescent Ratiometric Indicator for Analysis of Water Hardness in Household Water. Sensors 2020, 20, 3164. [Google Scholar] [CrossRef] [PubMed]
- Ameen, S.; Ahmad, M.; Mohsin, M.; Qureshi, M.I.; Ibrahim, M.M.; Abdin, M.Z.; Ahmad, A. Designing, construction and characterization of genetically encoded FRET-based nanosensor for real time monitoring of lysine flux in living cells. J. Nanobiotechnol. 2016, 14, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohsin, M.; Abdin, M.Z.; Nischal, L.; Kardam, H.; Ahmad, A. Genetically encoded FRET-based nanosensor for in vivo measurement of leucine. Biosens. Bioelectron. 2013, 50, 72–77. [Google Scholar] [CrossRef]
- Kausar, H.; Ambrin, G.; Okla, M.K.; Soufan, W.; Al-Ghamdi, A.A.; Ahmad, A. Metabolic Flux Analysis of Catechin Biosynthesis Pathways Using Nanosensor. Antioxidants 2020, 9, 288. [Google Scholar] [CrossRef] [Green Version]
- Kausar, H.; Ambrin, G.; Okla, M.K.; Alamri, S.A.; Soufan, W.H.; Ibrahim, E.I.; Abdel-Maksoud, M.A.; Ahmad, A. FRET-Based Genetically Encoded Nanosensor for Real-Time Monitoring of the Flux of α-Tocopherol in Living Cells. ACS Omega 2021, 6, 9020–9027. [Google Scholar] [CrossRef]
- Ambrin, G.; Ali, H.M.; Ahmad, A. Metabolic Regulation Analysis of Ajmalicine Biosynthesis Pathway in Catharanthus roseus (L.) G. Don Suspension Culture Using Nanosensor. Processes 2020, 8, 589. [Google Scholar] [CrossRef]
- Naz, R.; Okla, M.K.; Fatima, U.; Mohsin, M.; Soufan, W.H.; Alaraidh, I.A.; Abdel-Maksoud, M.A.; Ahmad, A. Designing and Development of FRET-Based Nanosensor for Real Time Analysis of N-Acetyl-5-Neuraminic Acid in Living Cells. Front. Nutr. 2021, 8, 621273. [Google Scholar] [CrossRef]
- Cho, K.F.; Javier, N.; Choi, K. BRET measurement on CCD camera-based microtiter plate readers. SLAS Discov. 2022, 27, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Weng, X.; Fu, Z.; Zhang, C.; Jiang, W.; Jiang, H. A Portable 3D Microfluidic Origami Biosensor for Cortisol Detection in Human Sweat. Anal. Chem. 2022, 94, 3526–3534. [Google Scholar] [CrossRef] [PubMed]
- Rutkauskaite, J.; Berger, S.; Stavrakis, S.; Dressler, O.; Heyman, J.; Casadevall i Solvas, X.; deMello, A.; Mazutis, L. High-throughput single-cell antibody secretion quantification and enrichment using droplet microfluidics-based FRET assay. iScience 2022, 25, 104515. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Oudeng, G.; Feng, H.; Liu, S.; Li, H.W.; Ho, Y.P.; Chen, Y.; Tan, Y.; Yang, M. 2D MOF Nanosensor-Integrated Digital Droplet Microfluidic Flow Cytometry for In Situ Detection of Multiple miRNAs in Single CTC Cells. Small 2022, 18, e2201779. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Yan, W.; Dong, F.; Hu, X.; Xu, Y.; Wang, Z.; Shen, Y.; Wang, W.; Zhao, Y.; Wei, W. A smartphone-based platform for ratiometric visualization of SARS-CoV-2 via an oligonucleotide probe. Mikrochim. Acta 2022, 189, 268. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Guo, Y.; Ma, Y.J.; Zhou, M.; Zhao, Y.Q.; Wang, J.F.; Fang, Y.J. Smartphone-assisted multiple-mode assay of ascorbic acid using cobalt oxyhydroxide nanoflakes and carbon quantum dots. Microchem. J. 2022, 175, 107185. [Google Scholar] [CrossRef]
- Lai, W.-Q.; Chang, Y.-F.; Chou, F.-N.; Yang, D.-M. Portable FRET-Based Biosensor Device for On-Site Lead Detection. Biosensors 2022, 12, 157. [Google Scholar] [CrossRef]
- Liu, T.; Chen, S.; Ruan, K.; Zhang, S.; He, K.; Li, J.; Chen, M.; Yin, J.; Sun, M.; Wang, X.; et al. A handheld multifunctional smartphone platform integrated with 3D printing portable device: On-site evaluation for glutathione and azodicarbonamide with machine learning. J. Hazard. Mater. 2022, 426, 128091. [Google Scholar] [CrossRef]


| System | Donor | Acceptor | Substrate | Features | Ref |
|---|---|---|---|---|---|
| BRET1 | Rluc/Rluc8 | EYFP | Coelenterazine-h | [39] | |
| BRET2 | Rluc | GFP2/GFP10 | DeepBlueC (bisdeoxycoelenterazine, coelenterazine 400a, di-dehydro coelenterazine) | Enlarged separation of donor and acceptor emission spectra, higher signal resolution | [40,41] |
| BRET3 | Rluc8 | mOrange | Coelenterazine-h | Longer wavelengths of the emission light and weaker attenuation of biological tissue | [41,45] |
| eBRET | Rluc | eYFP | EnduRen | Prolonged detection timescale from minutes to hours, enhanced luminescence intensity | [42,43,44] |
| BRET3.1 | Rluc8 | mOrange | Coelenterazine-v | [46] | |
| BRET4 | Rluc8 | TagRFP | Coelenterazine-h | [47] | |
| BRET4.1 | Rluc8 | TagRFP | Coelenterazine-v | [46] | |
| BRET5 | Rluc8.6 | TagRFP | Coelenterazine-h | [46] | |
| BRET6 | Rluc8.6 | TurboFP635 | Coelenterazine-h | [46] | |
| BRET6.1 | Rluc8.6 | TurboFP636 | Coelenterazine-v | [46] | |
| BRET7 | Rluc8 | TurboFP637 | Coelenterazine-v | [47] | |
| BRET8 | Rluc8.6 | TurboFP638 | Coelenterazine-h | [47] | |
| BRET9 | ALuc23 | FP, such as mCherry | Coelenterazine | A conceptually unique ligand-activatable BRET system | [48] |
| Principle | Analyte | Donor/Acceptor | Source of Sample | LOD/Linear Range | Ref. |
|---|---|---|---|---|---|
| FRET | Exosome | Cy3/MXenes | 1.4 × 103 particles mL−1 | [57] | |
| FRET | EpCAM | QDs/Cy3 | Serum | 600 pM | [58] |
| FRET | Dopamine | FAM/TAMRA | Serum | [59] | |
| FRET | Isoleucine | ECFP/Venus | Live cells | [60] | |
| FRET | Polysialic acid (PSA) | SQDs/Cy5 | Serum | 0.63 pM, 10 pM to 1 μM | [61] |
| FRET | cGMP | CFP/Venus, T-Sapphire/Dimer2 | Live cells | [62] | |
| FRET | Glutathione (GSH) and SO2 | CM/BP | Live cells | 75 μM for GSH and 0.16 μM for SO2 | [63] |
| BRET | Small molecule drugs | NanoLuc/Cy3 | Whole blood | [66] | |
| BRET | Antibodies such as those against HIV1-p17, hemagglutinin (HA), and dengue virus type I | NanoLuc/mNeonGreen | Plasma | 10 pM | [67,68,69] |
| BRET | Antibodies such as antiHIV1, anti-HA, and anti-DEN1 | NanoLuc/mNeonGreen | Whole blood | LODs of 2.8 nm, 7.1 nm, and 19.3 nm for anti-HIV1, anti-HA, and anti-DEN1, respectively | [70] |
| BRET | Antigen such as osteocalcin/BGP | NanoLuc/maleimide dye in Q-body | Solution | 0.11 nM | [72] |
| BRET | Metabolites | NanoLuc/Cy3 | Whole blood | [73] | |
| BRET | miRNA | NanoLuc/mNeonGreen | Serum | [74] |
| Principle | Analyte | Donor/Acceptor | Source of Samples | LOD/Linear Range | Ref. |
|---|---|---|---|---|---|
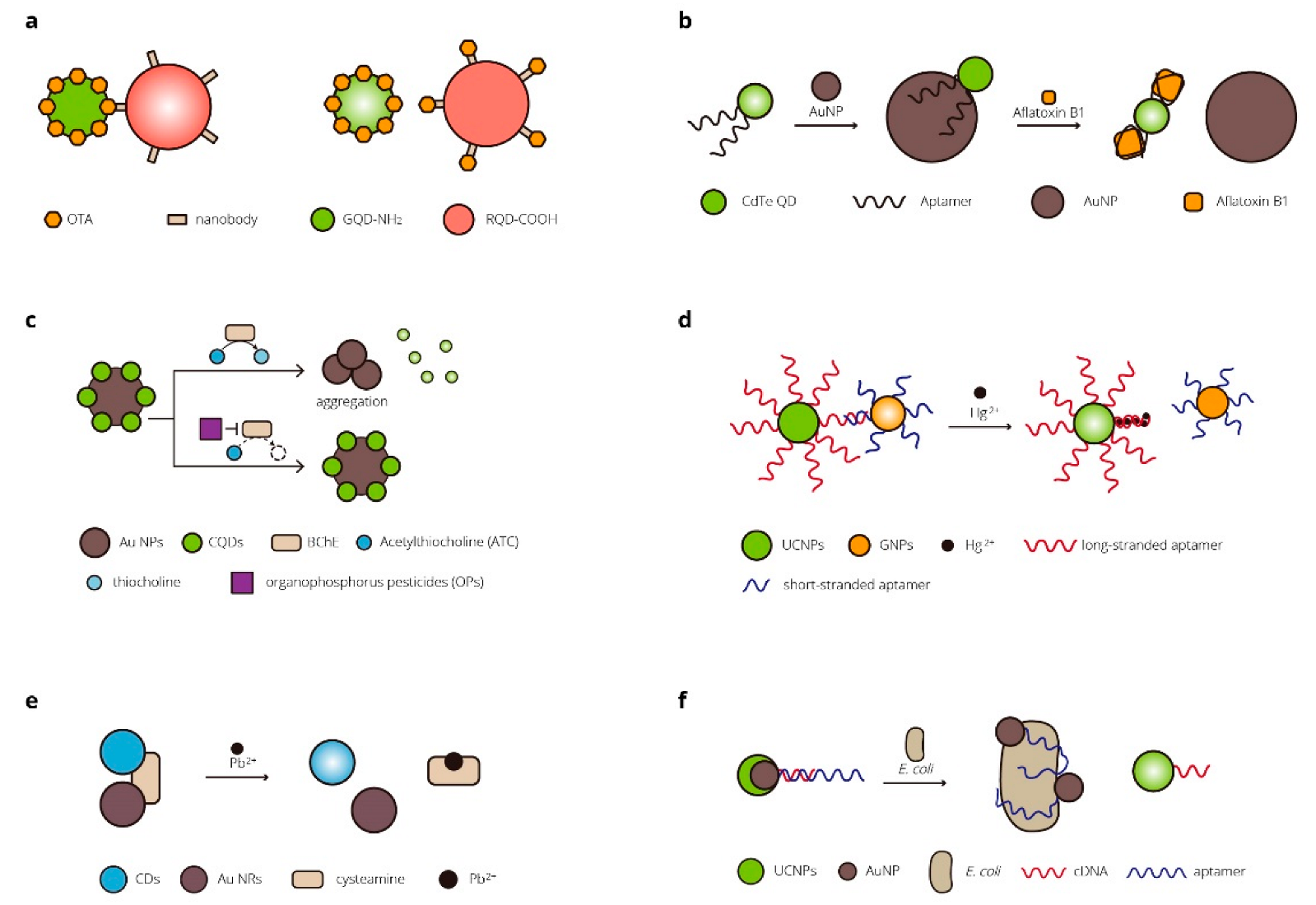
| FRET | Ochratoxin A (OTA) | QDs of different sizes | Agro-products | 5 pg/mL | [88] |
| FRET | Aflatoxin B1 (AFB1) | QDs/AuNPs | Agro-products | 3.4 nM, 10–400 nM | [89] |
| FRET | Organophosphorus pesticides (Ops) | CQDs/AuNPs | Tap and river water samples | 0.05 μg L−1, 0.05–50 μg L−1 | [90] |
| FRET | Hg2+ | UCNPs/GNPs | Tap water and milk samples | 60 nM, 0.2–20 μM | [91] |
| FRET | Pb2+ | CDs/Au NRs | Tap water and river water samples | 0.05 μM, 0 to 155 μM | [92] |
| FRET | Bacteria | UCNPs/AuNPs | Food and water samples | 3 cfu/mL, 5–106 cfu/mL | [93] |
| BRET | Ca2+/Mg2+ | NanoLuc/Venus | Water | [94] |
| Target Molecule | Donor/Acceptor | Sensor | Kd | LOD/Linear Range | Host | Ref. |
|---|---|---|---|---|---|---|
| Lysine | CFP and YFP | LAO | 97μM | Escherichia coli and Saccharomyces cerevisiae | [95] | |
| Leucine | CFP and YFP | LivK | 192 mM, 510 mM, 50 mM, and 105 mM, respectively, in different types | 900 mM, 10–1000 mM, 8.0–500 mM, and 150–800 Mm, respectively, in different types | Escherichia coli and S. cerevisiae | [96] |
| (+)-Catechin | ECFP and Venus | fraa-3 | 139 µM | Escherichia coli | [97] | |
| α-Tocopherol | ECFP and Venus | TTPA | 100 µM | Escherichia coli | [98] | |
| Ajmalicine | ECFP and Venus | CYP2D6 | Catharanthus roseus (L.) G. Don | [99] | ||
| N-acetyl-5-neuraminic acid (NeuAc) | ECFP and Venus | SiaP | ∼157 µM | Escherichia coli | [100] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Jiang, T. Developments in FRET- and BRET-Based Biosensors. Micromachines 2022, 13, 1789. https://doi.org/10.3390/mi13101789
Wu Y, Jiang T. Developments in FRET- and BRET-Based Biosensors. Micromachines. 2022; 13(10):1789. https://doi.org/10.3390/mi13101789
Chicago/Turabian StyleWu, Yuexin, and Tianyu Jiang. 2022. "Developments in FRET- and BRET-Based Biosensors" Micromachines 13, no. 10: 1789. https://doi.org/10.3390/mi13101789
APA StyleWu, Y., & Jiang, T. (2022). Developments in FRET- and BRET-Based Biosensors. Micromachines, 13(10), 1789. https://doi.org/10.3390/mi13101789





