Rapid Detection of Malaria Based on Hairpin-Mediated Amplification and Lateral Flow Detection
Abstract
:1. Introduction
2. Experimental Methods
2.1. Materials and Reagents
2.2. HMA-LFD Protocol
2.3. Performance Testing of HMA-LFD
3. Results
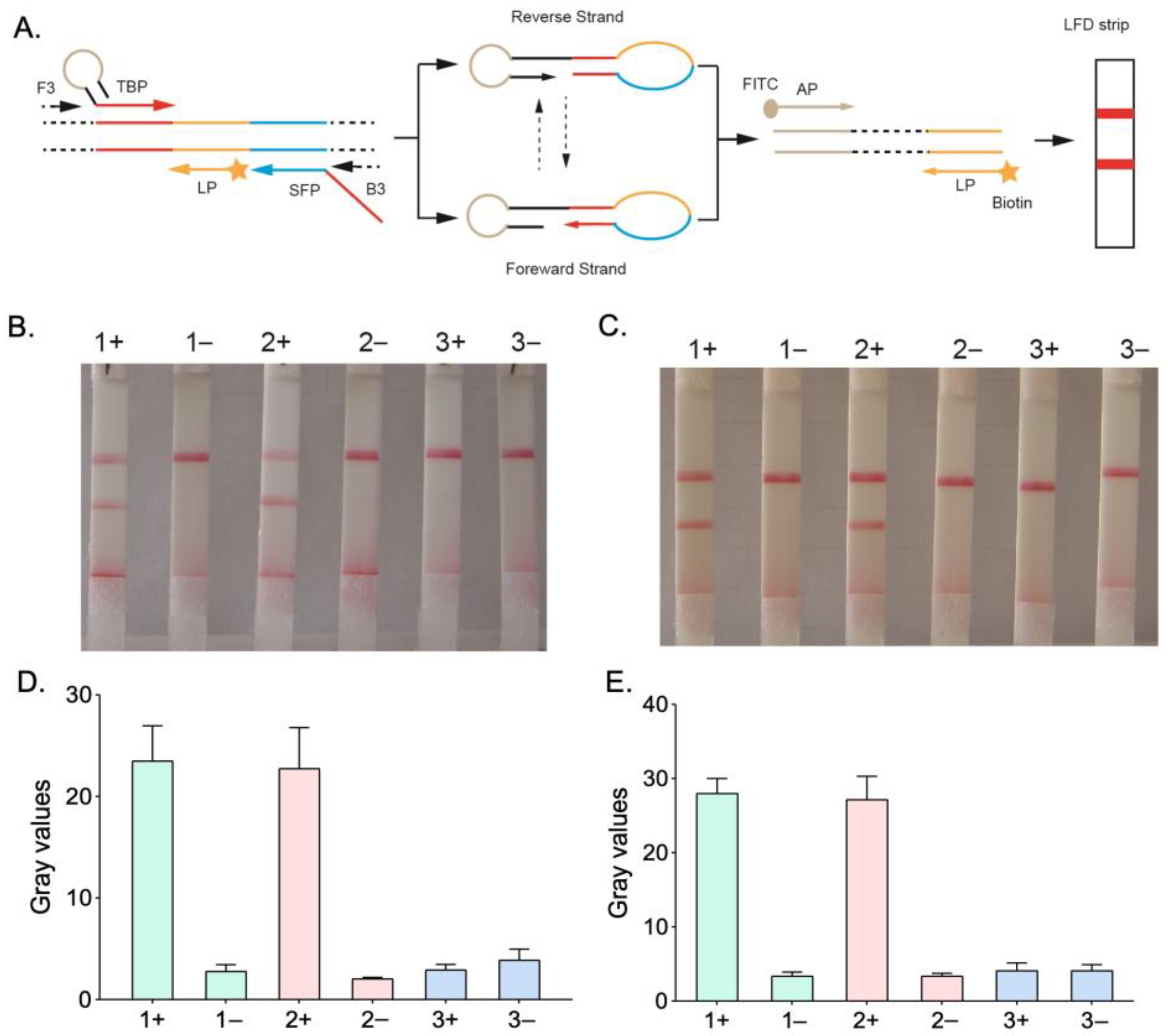
3.1. Optimization of the HMA-LFD System
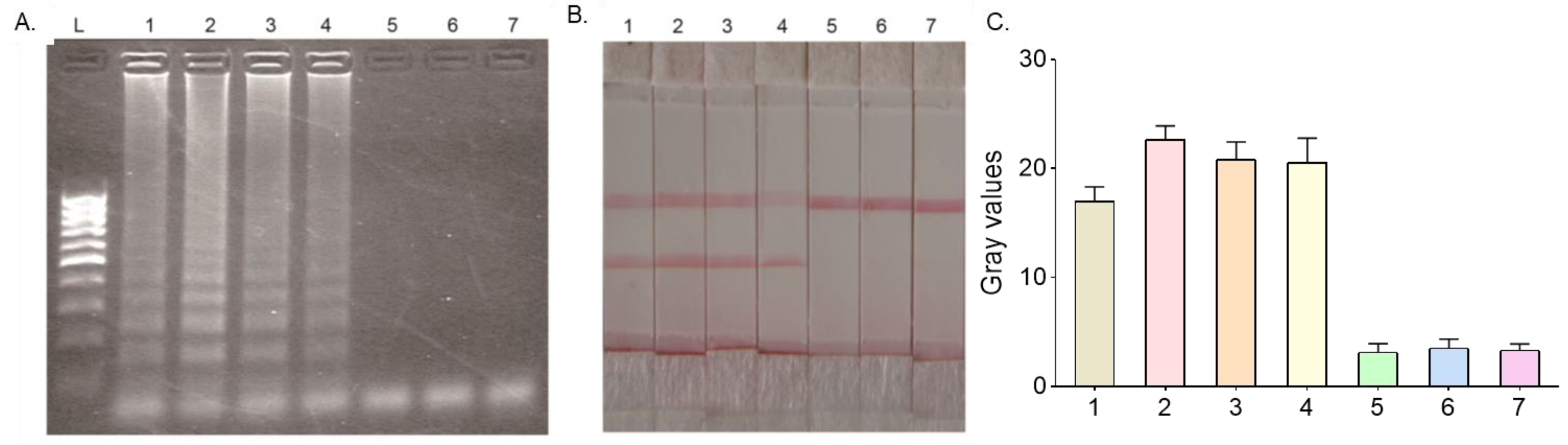
3.2. Specificity of HMA-LFD Detection System
3.3. Sensitivity of HMA-LFD Detection System
3.4. Validation of the HMA-LFD Detection System Using Clinical Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Phyo, A.P.; Dahal, P.; Mayxay, M.; Ashley, E.A. Clinical impact of vivax malaria: A collection review. PLoS Med. 2022, 19, e1003890. [Google Scholar] [CrossRef]
- World Malaria Report 2022. World Health Organization. Available online: https://www.who.int/publications/i/item/9789240064898 (accessed on 8 December 2022).
- Abeje, G.; Gelaye, W.; Alemu, G. Comparison of capillary, venous and buffy coat blood samples in detecting Plasmodium species among malaria suspected patients attending at Hamusite health center. A cross-sectional study. BMC Infect. Dis. 2021, 21, 576. [Google Scholar] [CrossRef]
- Kaewkamnerd, S.; Uthaipibull, C.; Intarapanich, A.; Pannarut, M.; Chaotheing, S.; Tongsima, S. An automatic device for detection and classification of malaria parasite species in thick blood film. BMC Bioinform. 2012, 13 (Suppl. 17), S18. [Google Scholar] [CrossRef]
- Cuadros, J.; Pérez-Tanoira, R.; Prieto-Pérez, L.; Martin-Martin, I.; Berzosa, P.; González, V.; Tisiano, G.; Balcha, S.; Ramos, J.M.; Górgolas, M. Field Evaluation of Malaria Microscopy, Rapid Malaria Tests and Loop-Mediated Isothermal Amplification in a Rural Hospital in South Western Ethiopia. PLoS ONE 2015, 10, e0142842. [Google Scholar] [CrossRef]
- Kotepui, M.; Uthaisar, K.; Phunphuech, B.; Phiwklam, N. A diagnostic tool for malaria based on computer software. Sci. Rep. 2015, 5, 16656. [Google Scholar] [CrossRef]
- Xu, G.; Nolder, D.; Reboud, J.; Oguike, M.C.; van Schalkwyk, D.A.; Sutherland, C.J.; Cooper, J.M. Paper-Origami-Based Multiplexed Malaria Diagnostics from Whole Blood. Angew. Chem. Int. Ed. Engl. 2016, 55, 15250–15253. [Google Scholar] [CrossRef]
- Hommel, B.; Charuel, J.-L.; Jaureguiberry, S.; Arnaud, L.; Courtin, R.; Kassab, P.; Prendki, V.; Paris, L.; Ghillani-Dalbin, P.; Thellier, M.; et al. Chronic malaria revealed by a new fluorescence pattern on the antinuclear autoantibodies test. PLoS ONE 2014, 9, e88548. [Google Scholar] [CrossRef]
- Zhang, C.-X.; Tanner, J.A.; Li, H.-W.; Wu, Y. A novel fluorescence probe of Plasmodium vivax lactate dehydrogenase based on adenosine monophosphate protected bimetallic nanoclusters. Talanta 2020, 213, 120850. [Google Scholar] [CrossRef]
- Lee, W.J.; Kim, H.H.; Hwang, S.M.; Park, M.Y.; Kim, N.R.; Cho, S.H. Detection of an antibody against Plasmodium vivax in residents of Gimpo-si, South Korea, using an indirect fluorescent antibody test. Malar. J. 2011, 10, 19. [Google Scholar] [CrossRef]
- DeSousa, J.M.; Jorge, M.Z.; Lindsay, H.B.; Haselton, F.R.; Wright, D.W.; Scherr, T.F. Inductively coupled plasma optical emission spectroscopy as a tool for evaluating lateral flow assays. Anal. Methods 2021, 13, 2137–2146. [Google Scholar] [CrossRef]
- Peng, Y.; Wu, J.; Wang, J.; Li, W.; Yu, S. Study and evaluation of Wondfo rapid diagnostic kit based on nano-gold immunochromatography assay for diagnosis of Plasmodium falciparum. Parasitol. Res. 2012, 110, 1421–1425. [Google Scholar] [CrossRef]
- Chiodini, P.L. Malaria diagnostics: Now and the future. Parasitology 2014, 141, 1873–1879. [Google Scholar] [CrossRef]
- Sazed, S.A.; Kibria, M.G.; Alam, M.S. An Optimized Real-Time qPCR Method for the Effective Detection of Human Malaria Infections. Diagnostics 2021, 11, 736. [Google Scholar] [CrossRef]
- Frickmann, H.; Weinreich, F.; Loderstädt, U.; Poppert, S.; Tannich, E.; Bull, J.; Kreikemeyer, B.; Barrantes, I. Metagenomic Sequencing for the Diagnosis of Plasmodium spp. with Different Levels of Parasitemia in EDTA Blood of Malaria Patients-A Proof-of-Principle Assessment. Int. J. Mol. Sci. 2022, 23, 11150. [Google Scholar] [CrossRef]
- O’flaherty, K.; Oo, W.H.; Zaloumis, S.G.; Cutts, J.C.; Aung, K.Z.; Thein, M.M.; Drew, D.R.; Razook, Z.; Barry, A.E.; Parischa, N.; et al. Community-based molecular and serological surveillance of subclinical malaria in Myanmar. BMC Med. 2021, 19, 121. [Google Scholar] [CrossRef]
- Lee, R.A.; Puig, H.D.; Nguyen, P.Q.; Angenent-Mari, N.M.; Donghia, N.M.; McGee, J.P.; Dvorin, J.D.; Klapperich, C.M.; Pollock, N.R.; Collins, J.J. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc. Natl. Acad. Sci. USA 2020, 117, 25722–25731. [Google Scholar] [CrossRef]
- McBirney, S.E.; Chen, D.; Scholtz, A.; Ameri, H.; Armani, A.M. Rapid Diagnostic for Point-of-Care Malaria Screening. ACS Sens. 2018, 3, 1264–1270. [Google Scholar] [CrossRef]
- Wang, C.; Liu, M.; Wang, Z.; Li, S.; Deng, Y.; He, N. Point-of-care diagnostics for infectious diseases: From methods to devices. Nano Today 2021, 37, 101092. [Google Scholar] [CrossRef]
- Li, H.; Steckl, A.J. Paper Microfluidics for Point-of-Care Blood-Based Analysis and Diagnostics. Anal. Chem. 2019, 91, 352–371. [Google Scholar] [CrossRef]
- Ruiz-Vega, G.; Arias-Alpízar, K.; de la Serna, E.; Borgheti-Cardoso, L.N.; Sulleiro, E.; Molina, I.; Fernàndez-Busquets, X.; Sánchez-Montalvá, A.; del Campo, F.J.; Baldrich, E. Electrochemical POC device for fast malaria quantitative diagnosis in whole blood by using magnetic beads, Poly-HRP and microfluidic paper electrodes. Biosens. Bioelectron. 2020, 150, 111925. [Google Scholar] [CrossRef]
- Shan, H.; Wang, Y.; Wu, T.; Ying, B.; Xu, G. Development of asymmetric hairpins-mediated nucleic acid isothermal amplification-based lateral flow detection of Mycobacterium tuberculosis. Sens. Actuators B Chem. 2022, 350, 130836. [Google Scholar] [CrossRef]
- Zheng, G.; Dai, J.; Wang, H.; Li, L.; Yuan, D.; Bai, S.; Song, X.; Zhao, Y. A hairpin-mediated nicking enzymatic signal amplification for nucleic acids detection. Talanta 2021, 225, 121991. [Google Scholar] [CrossRef]
- WS 259-2006; Diagnostic Criteria For Malaria. National Health Commission of the People’s Republic of China: Beijing, China, 2006.
- Shan, H.; Zhu, G.; Zhang, Y.; Ke, L.; Yang, X.; Qiao, A.; Wei, B.; Wang, Y.; Fan, Y.; Du, M. Multiplex PCR-ASE functionalized microfluidic diagnostic platform for the detection of clarithromycin resistance mutations in Helicobacter pylori. Sens. Actuators B Chem. 2023, 387, 133808. [Google Scholar] [CrossRef]
- Rosenbohm, J.M.; Klapperich, C.M.; Cabodi, M. Tunable Duplex Semiquantitative Detection of Nucleic Acids with a Visual Lateral Flow Immunoassay Readout. Anal. Chem. 2022, 94, 3956–3962. [Google Scholar] [CrossRef]
- Zheng, T.; Li, X.; Si, Y.; Wang, M.; Zhou, Y.; Yang, Y.; Liang, N.; Ying, B.; Wu, P. Specific lateral flow detection of isothermal nucleic acid amplicons for accurate point-of-care testing. Biosens. Bioelectron. 2023, 222, 114989. [Google Scholar] [CrossRef]
- Li, Y.; Kang, T.; Park, H.G. One-pot, ultrasensitive, and multiplex detection of SARS-CoV-2 genes utilizing self-priming hairpin-mediated isothermal amplification. Biosens. Bioelectron. 2023, 237, 115522. [Google Scholar] [CrossRef]
- Song, J.; Kim, H.Y.; Kim, S.; Jung, Y.; Park, H.G. Self-priming phosphorothioated hairpin-mediated isothermal amplification. Biosens. Bioelectron. 2021, 178, 113051. [Google Scholar] [CrossRef]
- Song, J.Y.; Jung, Y.; Lee, S.; Park, H.G. Self-Priming Hairpin-Utilized Isothermal Amplification Enabling Ultrasensitive Nucleic Acid Detection. Anal. Chem. 2020, 92, 10350–10356. [Google Scholar] [CrossRef]
- Bastaki, H.; Carter, J.; Marston, L.; Cassell, J.; Rait, G. Time delays in the diagnosis and treatment of malaria in non-endemic countries: A systematic review. Travel. Med. Infect. Dis. 2018, 21, 21–27. [Google Scholar] [CrossRef]
- Kim, H.Y.; Ahn, J.K.; Lee, C.Y.; Park, H.G. A hairpin probe-mediated isothermal amplification method to detect target nucleic acid. Anal. Chim. Acta 2020, 1114, 7–14. [Google Scholar] [CrossRef]
- Ye, X.; Fang, X.; Li, Y.; Wang, L.; Li, X.; Kong, J. Sequence-Specific Probe-Mediated Isothermal Amplification for the Single-Copy Sensitive Detection of Nucleic Acid. Anal. Chem. 2019, 91, 6738–6745. [Google Scholar] [CrossRef] [PubMed]
- Abdullah Al-Maskri, A.A.; Jin, G.; Li, Y.; Talap, J.; Almoiliqy, M.; Apu, C.; Zeng, S.; Zhou, Y.; Cai, S. A self-assembly amplification strategy for ultra-sensitive detection of microRNA based on phosphorothioated probes. Talanta 2022, 249, 123618. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wu, Q.; Chen, Y.; Liu, X.; Wang, F.; Zhou, X. Amplified MicroRNA Detection and Intracellular Imaging Based on an Autonomous and Catalytic Assembly of DNAzyme. ACS Sens. 2019, 4, 110–117. [Google Scholar] [CrossRef]
- Hopkins, H.; González, I.J.; Polley, S.D.; Angutoko, P.; Ategeka, J.; Asiimwe, C.; Agaba, B.; Kyabayinze, D.J.; Sutherland, C.J.; Perkins, M.D.; et al. Highly sensitive detection of malaria parasitemia in a malaria-endemic setting: Performance of a new loop-mediated isothermal amplification kit in a remote clinic in Uganda. J. Infect. Dis. 2013, 208, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Puri, M.; Brar, H.K.; Madan, E.; Srinivasan, R.; Rawat, K.; Gorthi, S.S.; Kumari, G.; Sah, R.; Ojha, S.B.; Panigrahi, S.; et al. Rapid diagnosis of Plasmodium falciparum malaria using a point-of-care loop-mediated isothermal amplification device. Front. Cell Infect. Microbiol. 2022, 12, 961832. [Google Scholar] [CrossRef]
- Park, J.S.; Kim, S.; Han, J.; Kim, J.H.; Park, K.S. Equipment-free, salt-mediated immobilization of nucleic acids for nucleic acid lateral flow assays. Sens. Actuators B Chem. 2022, 351, 130975. [Google Scholar] [CrossRef]
- Lin, C.; Liu, Z.; Fang, F.; Zhao, S.; Li, Y.; Xu, M.; Peng, Y.; Chen, H.; Yuan, F.; Zhang, W.; et al. Next-Generation Rapid and Ultrasensitive Lateral Flow Immunoassay for Detection of SARS-CoV-2 Variants. ACS Sens. 2023. [Google Scholar] [CrossRef]

| Primer | Sequence (5′ to 3′) |
|---|---|
| F3 | 5-TCGCTTCTAACGGTGAAC |
| B3 | 5-AATTGATAGTATCAGCTATCCATA |
| SFP | 5-GGTGGAACACATTGTTTCATTTGATCTCATTCCAATGGAACCT |
| TBP | 5-TAACCACAGCCAGGTTAGGTGCTCGTGGTTAGGTGGAACACATTGTTTCATT |
| AP | 5-FITC-TAACCACAGCCAGGTTAG |
| LP | 5-BIOTIN-TGGACGTAACCTCCAGGC |
| HMA-LFD | qPCR | |
|---|---|---|
| Positive | Negative | |
| Positive | 37 | 2 |
| Negative | 1 | 38 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Ke, L.; Sun, T.; Liu, Y.; Wei, B.; Du, M. Rapid Detection of Malaria Based on Hairpin-Mediated Amplification and Lateral Flow Detection. Micromachines 2023, 14, 1917. https://doi.org/10.3390/mi14101917
Zhang Y, Ke L, Sun T, Liu Y, Wei B, Du M. Rapid Detection of Malaria Based on Hairpin-Mediated Amplification and Lateral Flow Detection. Micromachines. 2023; 14(10):1917. https://doi.org/10.3390/mi14101917
Chicago/Turabian StyleZhang, Yang, Lihui Ke, Tao Sun, Yang Liu, Bo Wei, and Minghua Du. 2023. "Rapid Detection of Malaria Based on Hairpin-Mediated Amplification and Lateral Flow Detection" Micromachines 14, no. 10: 1917. https://doi.org/10.3390/mi14101917





