Precision Phenotypic Profiling and Capture of Circulating Tumor Cells via a Vertical Laminar Flow-Stacked Microfluidic Chip
Abstract
:1. Introduction
2. Materials and Methods
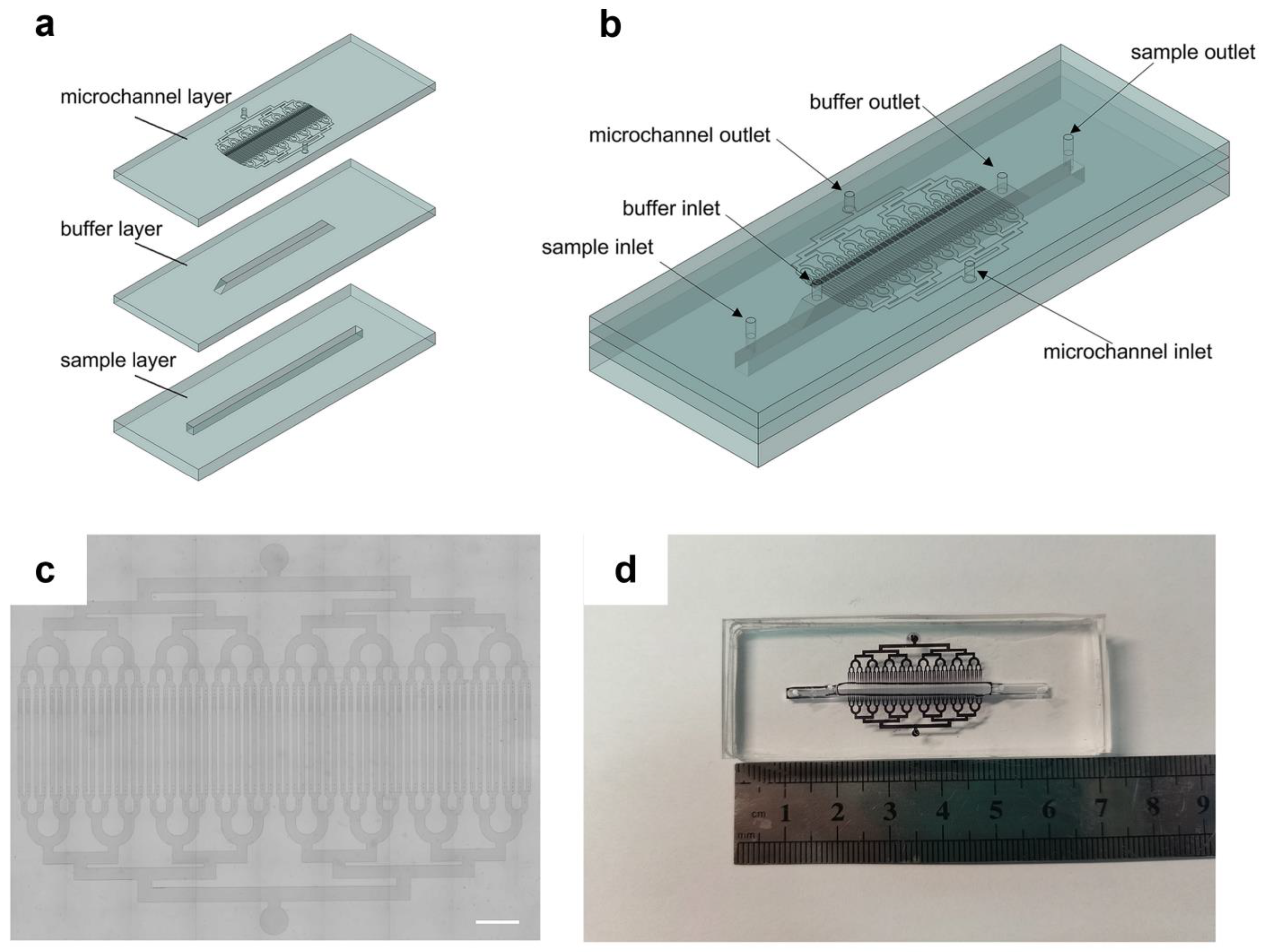
2.1. Device Design and Fabrication
2.2. Cell Culture and Experiment
2.3. Cell Labeling with EpCAM Immunomagnetic Beads
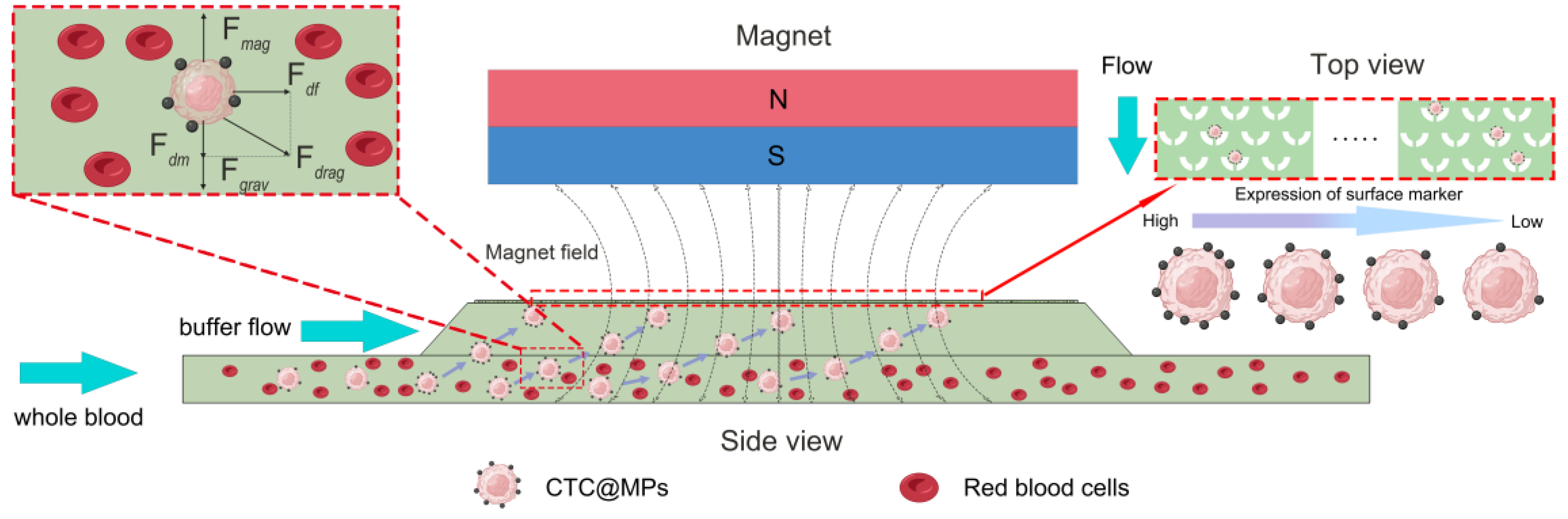
2.4. Preparation of Magnetic Field and Microfluidic Chip
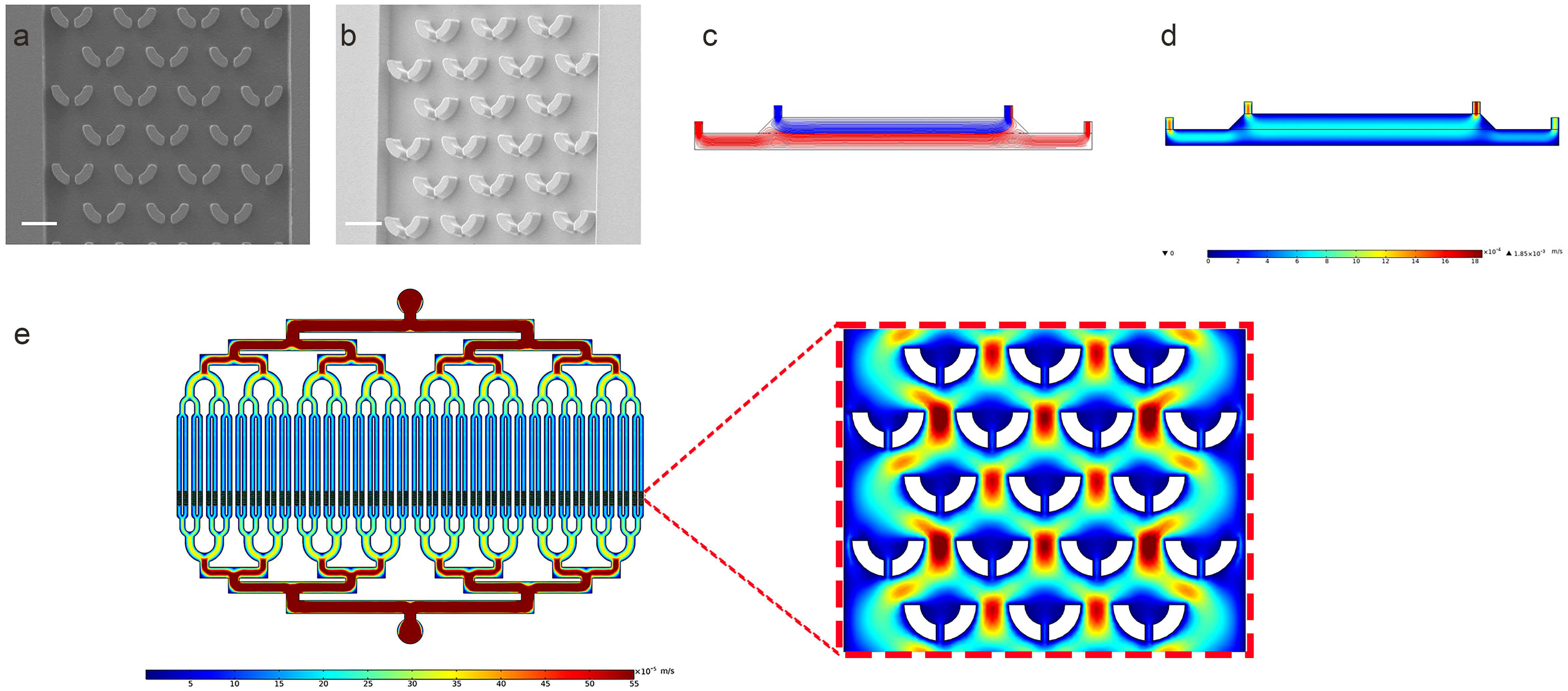
2.5. COMSOL Multiphysics Simulation
2.6. Spiking of Tumor Cells in Whole Blood
2.7. Quantitative Analysis of Cell Surface Expression
3. Results and Discussion
3.1. The Principle of Microfluidic Chips
3.2. Force Analysis of Cells
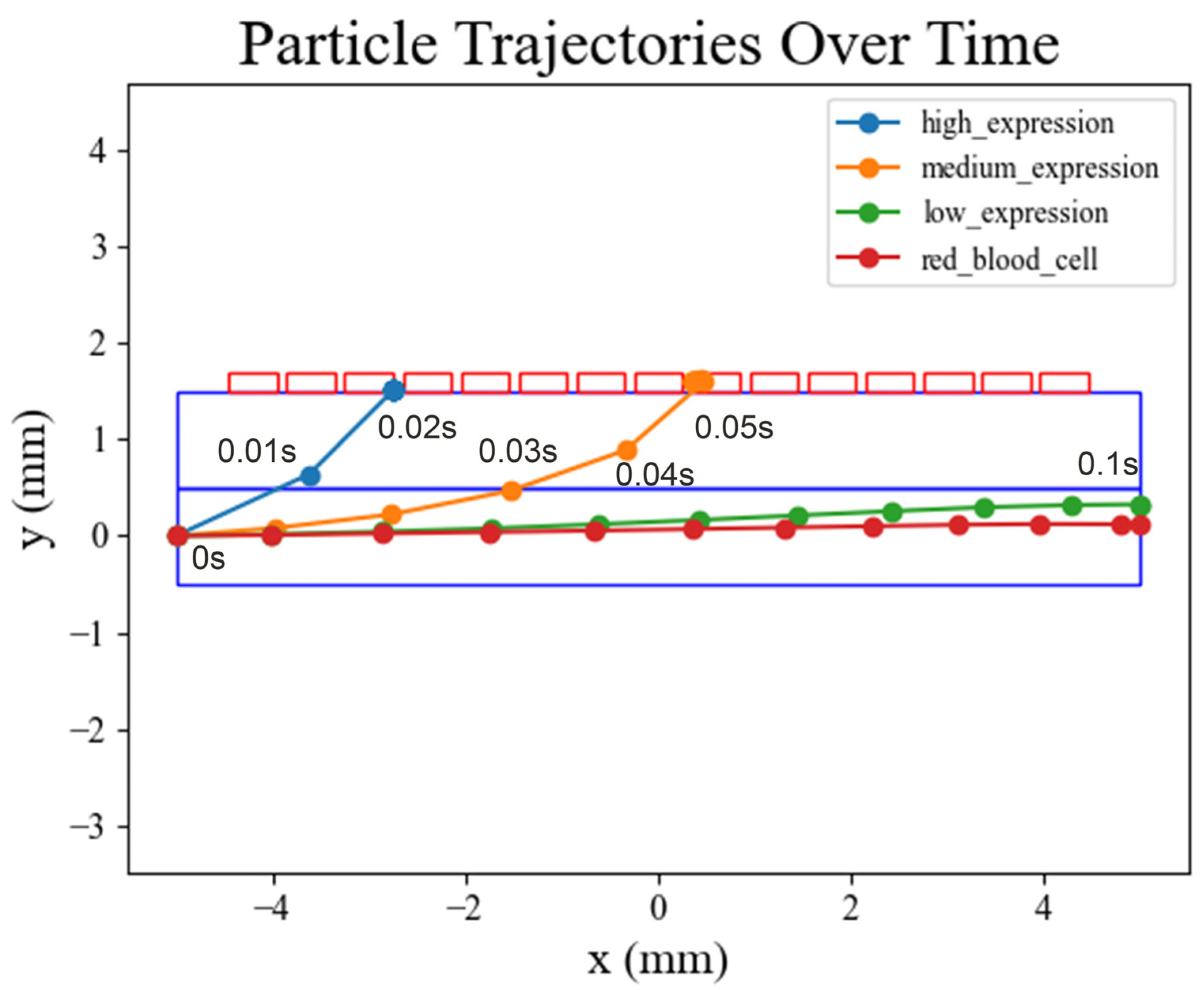
3.3. Cell Movement Trajectories in COMSOL Simulation
3.4. Characterization of the Microfluidic Chip
3.5. Cell Sorting and Capture Using Microfluidic Chip
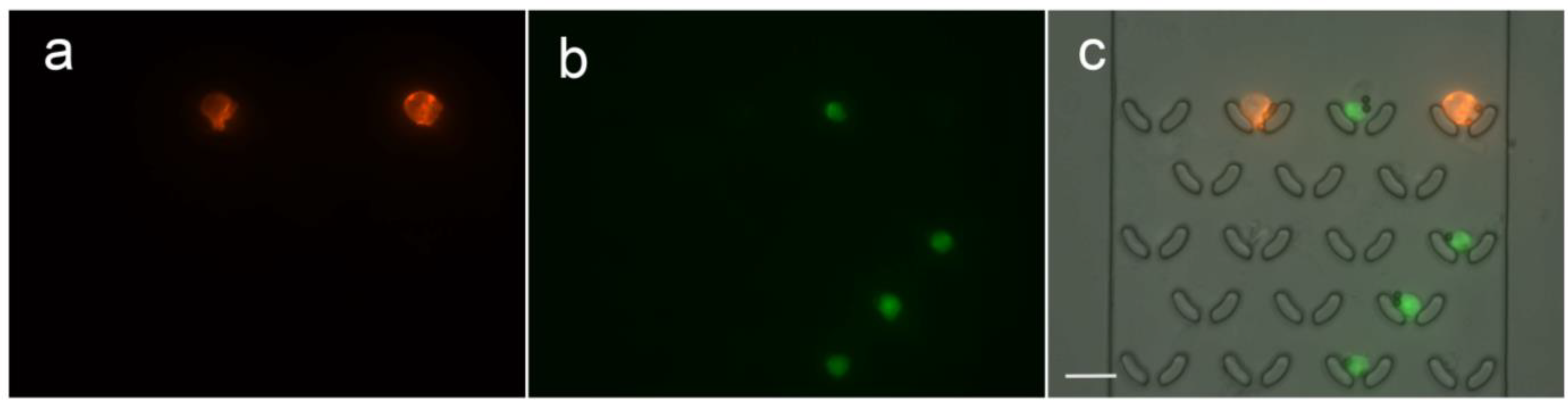
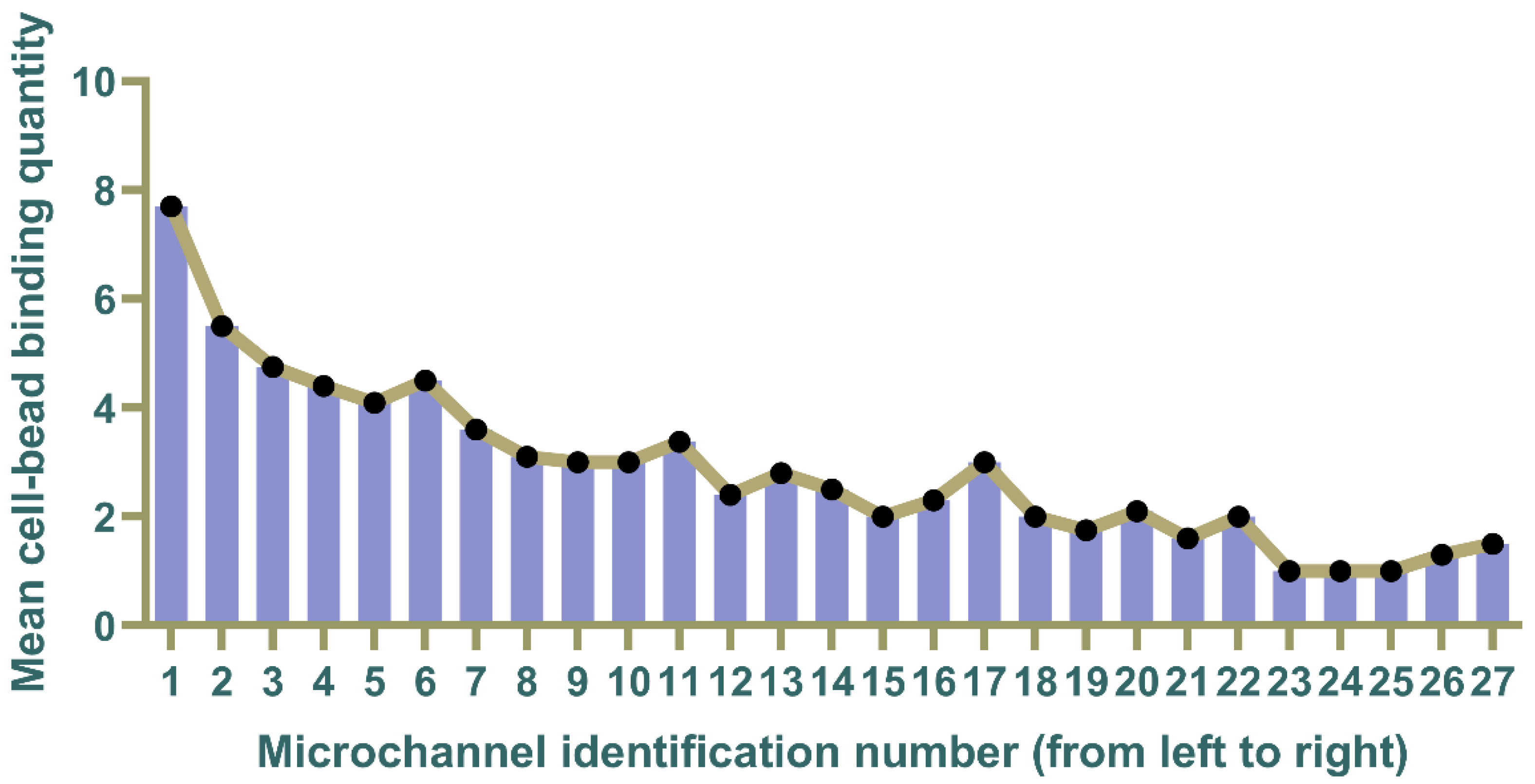
3.6. Quantification of Surface Expression Levels on Captured Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pantel, K.; Alix-Panabières, C.; Riethdorf, S. Cancer micrometastases. Nat. Rev. Clin. Oncol. 2009, 6, 339–351. [Google Scholar] [CrossRef]
- Nieto, M.A.; Huang, R.Y.-J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging biological principles of metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [PubMed]
- Poudineh, M.; Aldridge, P.M.; Ahmed, S.; Green, B.J.; Kermanshah, L.; Nguyen, V.; Tu, C.; Mohamadi, R.M.; Nam, R.K.; Hansen, A. Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking. Nat. Nanotechnol. 2017, 12, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Aya-Bonilla, C.A.; Morici, M.; Hong, X.; McEvoy, A.C.; Sullivan, R.J.; Freeman, J.; Calapre, L.; Khattak, M.A.; Meniawy, T.; Millward, M. Detection and prognostic role of heterogeneous populations of melanoma circulating tumour cells. Br. J. Cancer 2020, 122, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
- Ju, S.; Chen, C.; Zhang, J.; Xu, L.; Zhang, X.; Li, Z.; Chen, Y.; Zhou, J.; Ji, F.; Wang, L. Detection of circulating tumor cells: Opportunities and challenges. Biomark. Res. 2022, 10, 58. [Google Scholar] [CrossRef] [PubMed]
- Davnall, F.; Yip, C.S.; Ljungqvist, G.; Selmi, M.; Ng, F.; Sanghera, B.; Ganeshan, B.; Miles, K.A.; Cook, G.J.; Goh, V. Assessment of tumor heterogeneity: An emerging imaging tool for clinical practice? Insights Into Imaging 2012, 3, 573–589. [Google Scholar] [CrossRef]
- Just, N. Improving tumour heterogeneity MRI assessment with histograms. Br. J. Cancer 2014, 111, 2205–2213. [Google Scholar] [CrossRef]
- Alic, L.; Niessen, W.J.; Veenland, J.F. Quantification of heterogeneity as a biomarker in tumor imaging: A systematic review. PLoS ONE 2014, 9, e110300. [Google Scholar] [CrossRef]
- Jamal-Hanjani, M.; Quezada, S.A.; Larkin, J.; Swanton, C. Translational implications of tumor heterogeneity. Clin. Cancer Res. 2015, 21, 1258–1266. [Google Scholar] [CrossRef]
- Zhang, A.; Miao, K.; Sun, H.; Deng, C.-X. Tumor heterogeneity reshapes the tumor microenvironment to influence drug resistance. Int. J. Biol. Sci. 2022, 18, 3019. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Shen, L.; Luo, M.; Zhang, K.; Li, J.; Yang, Q.; Zhu, F.; Zhou, D.; Zheng, S.; Chen, Y. Circulating tumor cells: Biology and clinical significance. Signal Transduct. Target. Ther. 2021, 6, 404. [Google Scholar] [CrossRef] [PubMed]
- Harouaka, R.; Kang, Z.; Zheng, S.-Y.; Cao, L. Circulating tumor cells: Advances in isolation and analysis, and challenges for clinical applications. Pharmacol. Ther. 2014, 141, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Gertler, R.; Rosenberg, R.; Fuehrer, K.; Dahm, M.; Nekarda, H.; Siewert, J.R. Detection of circulating tumor cells in blood using an optimized density gradient centrifugation. In Molecular Staging of Cancer; Springer: Berlin/Heidelberg, Germany, 2003; pp. 149–155. [Google Scholar]
- Jørgensen, M.; Müller, C.; Sikkersoq, M.; Nadzieja, M.; Zhang, Z.; Su, Y.; Just, J.; Spindler, K.-L.G.; Chen, M. A melt-electrowritten filter for capture and culture of circulating colon cancer cells. Mater. Today Bio 2020, 6, 100052. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Lin, H.K.; Cote, R.J.; Tai, Y.C. A novel 3D micro membrane filtration device for capture viable rare circulating tumor cells from whole blood. In Proceedings of the Solid-State Sensors, Actuators, and Microsystems Workshop, Hilton Head Island, SC, USA, 1–5 June 2008; pp. 134–137. [Google Scholar]
- Stott, S.L.; Hsu, C.-H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.Y.; Hyun, K.-A.; Kim, S.-I.; Jung, H.-I. An integrated microfluidic chip for one-step isolation of circulating tumor cells. Sens. Actuators B Chem. 2017, 238, 1144–1150. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.-T.; Zhao, L.; Shen, Q.; Garcia, M.A.; Wu, D.; Hou, S.; Song, M.; Xu, X.; OuYang, W.-H.; OuYang, W.W.-L. NanoVelcro Chip for CTC enumeration in prostate cancer patients. Methods 2013, 64, 144–152. [Google Scholar] [CrossRef]
- Geens, M.; Van de Velde, H.; De Block, G.; Goossens, E.; Van Steirteghem, A.; Tournaye, H. The efficiency of magnetic-activated cell sorting and fluorescence-activated cell sorting in the decontamination of testicular cell suspensions in cancer patients. Hum. Reprod. 2007, 22, 733–742. [Google Scholar] [CrossRef]
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239. [Google Scholar] [CrossRef]
- Adams, A.A.; Okagbare, P.I.; Feng, J.; Hupert, M.L.; Patterson, D.; Göttert, J.; McCarley, R.L.; Nikitopoulos, D.; Murphy, M.C.; Soper, S.A. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J. Am. Chem. Soc. 2008, 130, 8633–8641. [Google Scholar] [CrossRef]
- Reátegui, E.; Aceto, N.; Lim, E.J.; Sullivan, J.P.; Jensen, A.E.; Zeinali, M.; Martel, J.M.; Aranyosi, A.J.; Li, W.; Castleberry, S. Tunable nanostructured coating for the capture and selective release of viable circulating tumor cells. Adv. Mater. 2015, 27, 1593–1599. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; Wong, I.Y.; Yanik, A.A.; Deen, W.M.; Toner, M. Discontinuous Nanoporous Membranes Reduce Non-Specific Fouling for Immunoaffinity Cell Capture. Small 2013, 9, 4207–4214. [Google Scholar] [CrossRef]
- Hosokawa, M.; Hayata, T.; Fukuda, Y.; Arakaki, A.; Yoshino, T.; Tanaka, T.; Matsunaga, T. Size-selective microcavity array for rapid and efficient detection of circulating tumor cells. Anal. Chem. 2010, 82, 6629–6635. [Google Scholar] [CrossRef] [PubMed]
- Gascoyne, P.R.; Noshari, J.; Anderson, T.J.; Becker, F.F. Isolation of rare cells from cell mixtures by dielectrophoresis. Electrophoresis 2009, 30, 1388–1398. [Google Scholar] [CrossRef]
- Shim, S.; Stemke-Hale, K.; Tsimberidou, A.M.; Noshari, J.; Anderson, T.E.; Gascoyne, P.R. Antibody-independent isolation of circulating tumor cells by continuous-flow dielectrophoresis. Biomicrofluidics 2013, 7, 011807. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Bessette, P.H.; Qian, J.; Meinhart, C.D.; Daugherty, P.S.; Soh, H.T. Marker-specific sorting of rare cells using dielectrophoresis. Proc. Natl. Acad. Sci. USA 2005, 102, 15757–15761. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, K.; Huang, Y.-Y.; Lane, N.; Huebschman, M.; Uhr, J.W.; Frenkel, E.P.; Zhang, X. Microchip-based immunomagnetic detection of circulating tumor cells. Lab Chip 2011, 11, 3449–3457. [Google Scholar] [CrossRef]
- Ozkumur, E.; Shah, A.M.; Ciciliano, J.C.; Emmink, B.L.; Miyamoto, D.T.; Brachtel, E.; Yu, M.; Chen, P.-i.; Morgan, B.; Trautwein, J. Inertial focusing for tumor antigen–dependent and–independent sorting of rare circulating tumor cells. Sci. Transl. Med. 2013, 5, 179ra147. [Google Scholar] [CrossRef]
- Talasaz, A.H.; Powell, A.A.; Huber, D.E.; Berbee, J.G.; Roh, K.-H.; Yu, W.; Xiao, W.; Davis, M.M.; Pease, R.F.; Mindrinos, M.N. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc. Natl. Acad. Sci. USA 2009, 106, 3970–3975. [Google Scholar] [CrossRef]
- Zhao, M.; Nelson, W.C.; Wei, B.; Schiro, P.G.; Hakimi, B.M.; Johnson, E.S.; Anand, R.K.; Gyurkey, G.S.; White, L.M.; Whiting, S.H. New generation of ensemble-decision aliquot ranking based on simplified microfluidic components for large-capacity trapping of circulating tumor cells. Anal. Chem. 2013, 85, 9671–9677. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.K.; McFaul, S.M.; Jin, C.; Black, P.C.; Ma, H. Highly selective biomechanical separation of cancer cells from leukocytes using microfluidic ratchets and hydrodynamic concentrator. Biomicrofluidics 2013, 7, 034114. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Yoon, H.J.; Stella, P.; Nagrath, S. Cascaded spiral microfluidic device for deterministic and high purity continuous separation of circulating tumor cells. Biomicrofluidics 2014, 8, 064117. [Google Scholar] [CrossRef] [PubMed]
- Fachin, F.; Spuhler, P.; Martel-Foley, J.M.; Edd, J.F.; Barber, T.A.; Walsh, J.; Karabacak, M.; Pai, V.; Yu, M.; Smith, K. Monolithic chip for high-throughput blood cell depletion to sort rare circulating tumor cells. Sci. Rep. 2017, 7, 10936. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, M.; Yamada, M.; Mitamura, R.; Ike, K.; Toyama, K.; Seki, M. Magnetophoresis-integrated hydrodynamic filtration system for size-and surface marker-based two-dimensional cell sorting. Anal. Chem. 2013, 85, 7666–7673. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.C.; Duffy, D.C.; Anderson, J.R.; Chiu, D.T.; Wu, H.; Schueller, O.J.; Whitesides, G.M. Fabrication of microfluidic systems in poly (dimethylsiloxane). Electrophor. Int. J. 2000, 21, 27–40. [Google Scholar] [CrossRef]
- Furlani, E. Analysis of particle transport in a magnetophoretic microsystem. J. Appl. Phys. 2006, 99, 024912. [Google Scholar] [CrossRef]
- Nam-Trung, N. Micro-magnetofluidics: Interactions between magnetism and fluid flow on the microscale. Microfluid. Nanofluid. 2012, 12, 1–16. [Google Scholar]
- Gijs, M.A.; Lacharme, F.; Lehmann, U. Microfluidic applications of magnetic particles for biological analysis and catalysis. Chem. Rev. 2010, 110, 1518–1563. [Google Scholar] [CrossRef]
- Ni, J.; Cozzi, P.J.; Duan, W.; Shigdar, S.; Graham, P.H.; John, K.H.; Li, Y. Role of the EpCAM (CD326) in prostate cancer metastasis and progression. Cancer Metastasis Rev. 2012, 31, 779–791. [Google Scholar] [CrossRef]
- Kimura, H.; Kato, H.; Faried, A.; Sohda, M.; Nakajima, M.; Fukai, Y.; Miyazaki, T.; Masuda, N.; Fukuchi, M.; Kuwano, H. Prognostic significance of EpCAM expression in human esophageal cancer. Int. J. Oncol. 2007, 30, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Tayama, S.; Motohara, T.; Narantuya, D.; Li, C.; Fujimoto, K.; Sakaguchi, I.; Tashiro, H.; Saya, H.; Nagano, O.; Katabuchi, H. The impact of EpCAM expression on response to chemotherapy and clinical outcomes in patients with epithelial ovarian cancer. Oncotarget 2017, 8, 44312. [Google Scholar] [CrossRef] [PubMed]
- Wen, K.-C.; Sung, P.-L.; Chou, Y.-T.; Pan, C.-M.; Wang, P.-H.; Lee, O.K.-S.; Wu, C.-W. The role of EpCAM in tumor progression and the clinical prognosis of endometrial carcinoma. Gynecol. Oncol. 2018, 148, 383–392. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Ma, Y.; Wang, Y.; Liang, Z.; Zhang, X.; Chen, Y.; Wang, Q.; Qin, H.; Wang, J. Precision Phenotypic Profiling and Capture of Circulating Tumor Cells via a Vertical Laminar Flow-Stacked Microfluidic Chip. Micromachines 2024, 15, 542. https://doi.org/10.3390/mi15040542
Zhang X, Ma Y, Wang Y, Liang Z, Zhang X, Chen Y, Wang Q, Qin H, Wang J. Precision Phenotypic Profiling and Capture of Circulating Tumor Cells via a Vertical Laminar Flow-Stacked Microfluidic Chip. Micromachines. 2024; 15(4):542. https://doi.org/10.3390/mi15040542
Chicago/Turabian StyleZhang, Xinping, Yuan Ma, Yujiao Wang, Zhenwei Liang, Xuanhe Zhang, Yiqing Chen, Qingyi Wang, Hua Qin, and Jiadao Wang. 2024. "Precision Phenotypic Profiling and Capture of Circulating Tumor Cells via a Vertical Laminar Flow-Stacked Microfluidic Chip" Micromachines 15, no. 4: 542. https://doi.org/10.3390/mi15040542




