Uniform Cell Distribution Achieved by Using Cell Deformation in a Micropillar Array
Abstract
:1. Introduction
2. Experimental Section
2.1. Principle
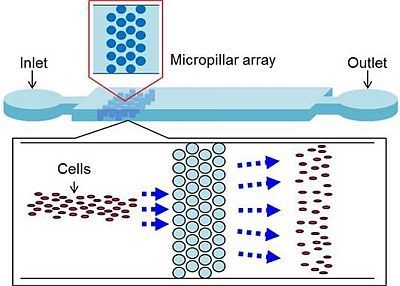
2.1.1. Uniform Cell Distribution by Micropillar Array




2.2. Experimental
2.2.1. Device Design

2.2.2. Device Fabrication

2.2.3. Cell Culture and Preparation of Cell Suspensions
2.2.4. Experimental Setup

3. Results and Discussion
3.1. Cell Introduction into Microchamber without Micropillar Array

3.2. Cell Introduction into Microchamber with Micropillar Array

3.3. Detailed Motion of N87 Cells in Micropillar Array



3.4. Comparison of Cell Distributions


4. Conclusions
Acknowledgments
Author Contributions
Supplementary Materials
Conflicts of Interest
References
- Andersson, H.; van den Berg, A. Microfluidic devices for cellomics: A review. Sens. Actuators B Chem. 2003, 92, 315–325. [Google Scholar] [CrossRef]
- Kim, S.M.; Lee, S.H.; Suh, K.Y. Cell research with physically modified microfluidic channels: A review. Lab Chip 2008, 8, 1015–1023. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, W.; Wang, Y.; Wang, J.; Tu, Q.; Liu, R.; Wang, J. Construction of oxygen and chemical concentration gradients in a single microfluidic device for studying tumor cell-drug interactions in a dynamic hypoxia microenvironment. Lab Chip 2013, 13, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A.; et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Marshall, D. Microfluidics for single cell analysis. Curr. Opin. Biotechnol. 2012, 23, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Zervantonakis, I.K.; Hughes-Alford, S.K.; Charest, J.L.; Condeelis, J.S.; Gertler, F.B.; Kamm, R.D. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl. Acad. Sci. USA 2012, 109, 13515–13520. [Google Scholar] [CrossRef] [PubMed]
- Squires, T.; Quake, S. Microfluidics Fluid physics at the nanoliter scale. Rev. Mod. Phys. 2005, 77, 977–1026. [Google Scholar] [CrossRef]
- Lee, P.J.; Hung, P.J.; Rao, V.M.; Lee, L.P. Nanoliter scale microbioreactor array for quantitative cell biology. Biotechnol. Bioeng. 2006, 94, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Brewer, G.J.; Torricelli, J.R.; Evege, E.K.; Price, P.J. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J. Neurosci. Res. 1993, 35, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Hui, T.Y.; Cheung, K.M.C.; Cheung, W.L.; Chan, D.; Chan, B.P. In vitro chondrogenic differentiation of human mesenchymal stem cells in collagen microspheres: Influence of cell seeding density and collagen concentration. Biomaterials 2008, 29, 3201–3212. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Wang, Y.; Lo, A.S.-Y.; Gomes, E.M.; Junghans, R.P. Cell density plays a critical role in ex vivo expansion of T cells for adoptive immunotherapy. J. Biomed. Biotechnol. 2010, 2010, 386545. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Thibeault, S. Effect of DMSO concentration, cell density and needle gauge on the viability of cryopreserved cells in three dimensional hyaluronan hydrogel. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2013, 2013, 6228–6231. [Google Scholar] [PubMed]
- Dar, A.; Shachar, M.; Leor, J.; Cohen, S. Optimization of cardiac cell seeding and distribution in 3D porous alginate scaffolds. Biotechnol. Bioeng. 2002, 80, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, J.; Pye, E.K. Cell density dependence of oscillatory metabolism. Nature 1976, 259, 670–671. [Google Scholar] [CrossRef] [PubMed]
- Kuszynski, C.A.; Miller, K.A.; Rizzino, A. Influence of cell density and receptor number on the binding and distribution of cell surface epidermal growth factor receptors. In Vitro Cell. Dev. Biol. Anim. 1993, 29A, 708–713. [Google Scholar] [CrossRef]
- Basu, S.; Gerchman, Y.; Collins, C.H.; Arnold, F.H.; Weiss, R. A synthetic multicellular system for programmed pattern formation. Nature 2005, 434, 1130–1134. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ze’ev, A.; Robinson, G.S.; Bucher, N.L.; Farmer, S.R. Cell-cell and cell-matrix interactions differentially regulate the expression of hepatic and cytoskeletal genes in primary cultures of rat hepatocytes. Proc. Natl. Acad. Sci. USA 1988, 85, 2161–2165. [Google Scholar] [CrossRef] [PubMed]
- Altman, G.H.; Horan, R.L.; Martin, I.; Farhadi, J.; Stark, P.R.H.; Volloch, V.; Richmond, J.C.; Vunjak-Novakovic, G.; Kaplan, D.L. Cell differentiation by mechanical stress. FASEB J. 2002, 16, 270–272. [Google Scholar] [PubMed]
- Vunjak-Novakovic, G.; Altman, G.; Horan, R.; Kaplan, D.L. Tissue engineering of ligaments. Annu. Rev. Biomed. Eng. 2004, 6, 131–156. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Dean, D.; Mikos, A.G.; Fisher, J.P. Effect of initial cell seeding density on early osteogenic signal expression of rat bone marrow stromal cells cultured on cross-linked poly(propylene fumarate) disks. Biomacromolecules 2009, 10, 1810–1817. [Google Scholar] [CrossRef] [PubMed]
- Gage, B.K.; Webber, T.D.; Kieffer, T.J. Initial cell seeding density influences pancreatic endocrine development during in vitro differentiation of human embryonic stem cells. PLoS One 2013, 8, e82076. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Kim, M.-C.; Marquez, M.; Thorsen, T. High-density microfluidic arrays for cell cytotoxicity analysis. Lab Chip 2007, 7, 740–745. [Google Scholar] [CrossRef] [PubMed]
- Lichtner, R.B.; Schirrmacher, V. Cellular distribution and biological activity of epidermal growth factor receptors in A431 cells are influenced by cell-cell contact. J. Cell. Biochem. 1990, 144, 303–312. [Google Scholar]
- Melchels, F.P.W.; Barradas, A.M.C.; van Blitterswijk, C.A.; de Boer, J.; Feijen, J.; Grijpma, D.W. Effects of the architecture of tissue engineering scaffolds on cell seeding and culturing. Acta Biomater. 2010, 6, 4208–4217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Macia, L.; Morrin, A.; Smyth, M.R.; Killard, A.J. Advanced printing and deposition methodologies for the fabrication of biosensors and biodevices. Analyst 2010, 135, 845–867. [Google Scholar] [CrossRef] [PubMed]
- Khademhosseini, A.; Suh, K.Y.; Jon, S.; Eng, G.; Yeh, J.; Chen, G.J.; Langer, R. A soft lithographic approach to fabricate patterned microfluidic channels. Anal. Chem. 2004, 76, 3675–3681. [Google Scholar] [CrossRef] [PubMed]
- Rozkiewicz, D.I.; Kraan, Y.; Werten, M.W.T.; de Wolf, F.A.; Subramaniam, V.; Ravoo, B.J.; Reinhoudt, D.N. Covalent microcontact printing of proteins for cell patterning. Chemistry 2006, 12, 6290–6297. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Evander, M.; Hammarström, B.; Laurell, T. Review of cell and particle trapping in microfluidic systems. Anal. Chim. Acta 2009, 649, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Ges, I.A.; Brindley, R.L.; Currie, K.P.M.; Baudenbacher, F.J. A microfluidic platform for chemical stimulation and real time analysis of catecholamine secretion from neuroendocrine cells. Lab Chip 2013, 13, 4663–4673. [Google Scholar] [CrossRef] [PubMed]
- Brammer, K.S.; Choi, C.; Frandsen, C.J.; Oh, S.; Jin, S. Hydrophobic nanopillars initiate mesenchymal stem cell aggregation and osteo-differentiation. Acta Biomater. 2011, 7, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.M.; Jiang, Z.; Bastmeyer, M.; Lahann, J. Physical aspects of cell culture substrates: Topography, roughness, and elasticity. Small 2012, 8, 336–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, W.-H.; Takeuchi, S. A trap-and-release integrated microfluidic system for dynamic microarray applications. Proc. Natl. Acad. Sci. USA 2007, 104, 1146–1151. [Google Scholar] [CrossRef] [PubMed]
- Park, J.G.; Frucht, H.; LaRocca, R.V.; Bliss, D.P.; Kurita, Y.; Chen, T.R.; Henslee, J.G.; Trepel, J.B.; Jensen, R.T.; Johnson, B.E. Characteristics of cell lines established from human gastric carcinoma. Cancer Res. 1990, 50, 2773–2780. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaminaga, M.; Ishida, T.; Kadonosono, T.; Kizaka-Kondoh, S.; Omata, T. Uniform Cell Distribution Achieved by Using Cell Deformation in a Micropillar Array. Micromachines 2015, 6, 409-422. https://doi.org/10.3390/mi6040409
Kaminaga M, Ishida T, Kadonosono T, Kizaka-Kondoh S, Omata T. Uniform Cell Distribution Achieved by Using Cell Deformation in a Micropillar Array. Micromachines. 2015; 6(4):409-422. https://doi.org/10.3390/mi6040409
Chicago/Turabian StyleKaminaga, Maho, Tadashi Ishida, Tetsuya Kadonosono, Shinae Kizaka-Kondoh, and Toru Omata. 2015. "Uniform Cell Distribution Achieved by Using Cell Deformation in a Micropillar Array" Micromachines 6, no. 4: 409-422. https://doi.org/10.3390/mi6040409